A highly contiguous genome assembly for the California quail (Callipepla californica)
- PMID: 36763048
- PMCID: PMC10287149
- DOI: 10.1093/jhered/esad008
A highly contiguous genome assembly for the California quail (Callipepla californica)
Abstract
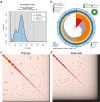
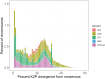
The California quail (Callipepla californica) is an iconic native bird of scrub and oak woodlands in California and the Baja Peninsula of Mexico. Here, we report a draft reference assembly for the species generated from PacBio HiFi long read and Omni-C chromatin-proximity sequencing data as part of the California Conservation Genomics Project (CCGP). Sequenced reads were assembled into 321 scaffolds totaling 1.08 Gb in length. Assembly metrics indicate a highly contiguous and complete assembly with a contig N50 of 5.5 Mb, scaffold N50 of 19.4 Mb, and BUSCO completeness score of 96.5%. Transposable elements (TEs) occupy 16.5% of the genome, more than previous Odontophoridae quail assemblies but in line with estimates of TE content for recent long-read assemblies of chicken and Peking duck. Together these metrics indicate that the present assembly is more complete than prior reference assemblies generated for Odontophoridae quail. This reference will serve as an essential resource for studies on local adaptation, phylogeography, and conservation genetics in this species of significant biological and recreational interest.
Keywords: California Conservation Genomics Project; Odontophoridae; transposable elements; upland game bird.
© The American Genetic Association. 2023.
Figures


References
-
- Bravo GA, Schmitt CJ, Edwards SV.. What have we learned from the first 500 avian genomes? Annu Rev Ecol Evol Syst. 2021;52(1):611–639. doi: 10.1146/annurev-ecolsys-012121-085928 - DOI
-
- Calkins JD, Gee JM, Hagelin JC, Lott DF.. California quail (Callipepla californica), version 1.0. In: Poole AF, editor. Birds of the world. Ithaca (NY): Cornell Lab of Ornithology; 2014.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

