Heterotrimeric G proteins regulate planarian regeneration and behavior
- PMID: 36763503
- PMCID: PMC10078920
- DOI: 10.1093/genetics/iyad019
Heterotrimeric G proteins regulate planarian regeneration and behavior
Abstract
G protein-coupled receptors play broad roles in development and stem cell biology, but few roles for G protein-coupled receptor signaling in complex tissue regeneration have been uncovered. Planarian flatworms robustly regenerate all tissues and provide a model with which to explore potential functions for G protein-coupled receptor signaling in somatic regeneration and pluripotent stem cell biology. As a first step toward exploring G protein-coupled receptor function in planarians, we investigated downstream signal transducers that work with G protein-coupled receptors, called heterotrimeric G proteins. Here, we characterized the complete heterotrimeric G protein complement in Schmidtea mediterranea for the first time and found that 7 heterotrimeric G protein subunits promote regeneration. We further characterized 2 subunits critical for regeneration, Gαq1 and Gβ1-4a, finding that they promote the late phase of anterior polarity reestablishment, likely through anterior pole-produced Follistatin. Incidentally, we also found that 5 G protein subunits modulate planarian behavior. We further identified a putative serotonin receptor, gcr052, that we propose works with Gαs2 and Gβx2 in planarian locomotion, demonstrating the utility of our strategy for identifying relevant G protein-coupled receptors. Our work provides foundational insight into roles of heterotrimeric G proteins in planarian biology and serves as a useful springboard toward broadening our understanding of G protein-coupled receptor signaling in adult tissue regeneration.
Keywords: Schmidtea; GPCR; behavior; flatworm; heterotrimeric G protein; neurobiology; planarian; regeneration; signaling.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Genetics Society of America.
Conflict of interest statement
Conflicts of interest None declared.
Figures


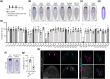


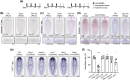

Similar articles
-
CBP/p300 homologs CBP2 and CBP3 play distinct roles in planarian stem cell function.Dev Biol. 2021 May;473:130-143. doi: 10.1016/j.ydbio.2021.02.004. Epub 2021 Feb 16. Dev Biol. 2021. PMID: 33607113
-
Cell-type diversity and regionalized gene expression in the planarian intestine.Elife. 2020 Apr 2;9:e52613. doi: 10.7554/eLife.52613. Elife. 2020. PMID: 32240093 Free PMC article.
-
Epigenetic regulation of planarian stem cells by the SET1/MLL family of histone methyltransferases.Epigenetics. 2013 Jan;8(1):79-91. doi: 10.4161/epi.23211. Epub 2012 Dec 12. Epigenetics. 2013. PMID: 23235145 Free PMC article.
-
The role of the EGFR signaling pathway in stem cell differentiation during planarian regeneration and homeostasis.Semin Cell Dev Biol. 2019 Mar;87:45-57. doi: 10.1016/j.semcdb.2018.05.011. Epub 2018 May 18. Semin Cell Dev Biol. 2019. PMID: 29775660 Review.
-
From worm to germ: Germ cell development and regeneration in planarians.Curr Top Dev Biol. 2019;135:127-153. doi: 10.1016/bs.ctdb.2019.04.001. Epub 2019 May 2. Curr Top Dev Biol. 2019. PMID: 31155357 Review.
Cited by
-
Ets-1 transcription factor regulates glial cell regeneration and function in planarians.Development. 2023 Sep 15;150(18):dev201666. doi: 10.1242/dev.201666. Epub 2023 Sep 15. Development. 2023. PMID: 37665145 Free PMC article.
-
A parenchymal niche regulates pluripotent stem cell function in planarians.bioRxiv [Preprint]. 2025 Aug 2:2025.08.01.668211. doi: 10.1101/2025.08.01.668211. bioRxiv. 2025. PMID: 40766428 Free PMC article. Preprint.
-
Combinatorial mechanisms specify cellular location and neurotransmitter identity during regeneration of planarian neurons.bioRxiv [Preprint]. 2025 May 25:2025.05.23.655781. doi: 10.1101/2025.05.23.655781. bioRxiv. 2025. PMID: 40661405 Free PMC article. Preprint.
-
The known, unknown, and unknown unknowns of cell-cell communication in planarian regeneration.Zool Res. 2023 Sep 18;44(5):981-992. doi: 10.24272/j.issn.2095-8137.2023.044. Zool Res. 2023. PMID: 37721107 Free PMC article. Review.
References
-
- Baguna J, Salo E, Auladell C. Regeneration and pattern formation in planarians. III. that neoblasts are totipotent stem cells and the cells. Development. 1989;107(1):77–86. doi:10.1242/dev.107.1.77. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

