Subnanometer structure of an enveloped virus fusion complex on viral surface reveals new entry mechanisms
- PMID: 36763666
- PMCID: PMC9917000
- DOI: 10.1126/sciadv.ade2727
Subnanometer structure of an enveloped virus fusion complex on viral surface reveals new entry mechanisms
Abstract
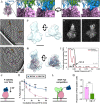
Paramyxoviruses-including important pathogens like parainfluenza, measles, and Nipah viruses-use a receptor binding protein [hemagglutinin-neuraminidase (HN) for parainfluenza] and a fusion protein (F), acting in a complex, to enter cells. We use cryo-electron tomography to visualize the fusion complex of human parainfluenza virus 3 (HN/F) on the surface of authentic clinical viruses at a subnanometer resolution sufficient to answer mechanistic questions. An HN loop inserts in a pocket on F, showing how the fusion complex remains in a ready but quiescent state until activation. The globular HN heads are rotated with respect to each other: one downward to contact F, and the other upward to grapple cellular receptors, demonstrating how HN/F performs distinct steps before F activation. This depiction of viral fusion illuminates potentially druggable targets for paramyxoviruses and sheds light on fusion processes that underpin wide-ranging biological processes but have not been visualized in situ or at the present resolution.
Figures




References
-
- M. Porotto, I. DeVito, S. G. Palmer, E. M. Jurgens, J. L. Yee, C. C. Yokoyama, A. Pessi, A. Moscona, Spring-loaded model revisited: Paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J. Virol. 85, 12867–12880 (2011). - PMC - PubMed
-
- M. Porotto, Z. W. Salah, L. Gui, I. DeVito, E. M. Jurgens, H. Lu, C. C. Yokoyama, L. M. Palermo, K. K. Lee, A. Moscona, Regulation of paramyxovirus fusion activation: The hemagglutinin-neuraminidase protein stabilizes the fusion protein in a pretriggered state. J. Virol. 86, 12838–12848 (2012). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

