LC-MS/MS-PRM Quantification of IgG Glycoforms Using Stable Isotope Labeled IgG1 Fc Glycopeptide Standard
- PMID: 36763792
- PMCID: PMC10461028
- DOI: 10.1021/acs.jproteome.2c00475
LC-MS/MS-PRM Quantification of IgG Glycoforms Using Stable Isotope Labeled IgG1 Fc Glycopeptide Standard
Abstract
Targeted quantification of proteins is a standard methodology with broad utility, but targeted quantification of glycoproteins has not reached its full potential. The lack of optimized workflows and isotopically labeled standards limits the acceptance of glycoproteomics quantification. In this work, we introduce an efficient and streamlined chemoenzymatic synthesis of a library of isotopically labeled glycopeptides of IgG1 which we use for quantification in an energy optimized LC-MS/MS-PRM workflow. Incorporation of the stable isotope labeled N-acetylglucosamine enables an efficient monitoring of all major fragment ions of the glycopeptides generated under the soft higher-energy C-trap dissociation (HCD) conditions, which reduces the coefficients of variability (CVs) of the quantification to 0.7-2.8%. Our results document, for the first time, that the workflow using a combination of stable isotope labeled standards with intrascan normalization enables quantification of the glycopeptides by an electron transfer dissociation (ETD) workflow, as well as the HCD workflow, with the highest sensitivity compared to traditional workflows. This was exemplified by a rapid quantification (13 min) of IgG1 Fc glycoforms from COVID-19 patients.
Keywords: Glycopeptide Synthesis; Glycoproteomics; Immunoglobulins; Mass Spectrometry; PRM Analysis.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
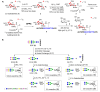
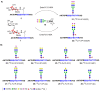
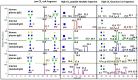


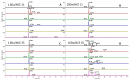
Update of
-
LC-MS/MS-PRM Quantification of IgG glycoforms using stable isotope labeled IgG1 Fc glycopeptide standard.bioRxiv [Preprint]. 2022 Aug 2:2022.08.02.501850. doi: 10.1101/2022.08.02.501850. bioRxiv. 2022. Update in: J Proteome Res. 2023 Apr 7;22(4):1138-1147. doi: 10.1021/acs.jproteome.2c00475. PMID: 35982648 Free PMC article. Updated. Preprint.
Similar articles
-
LC-MS/MS-PRM Quantification of IgG glycoforms using stable isotope labeled IgG1 Fc glycopeptide standard.bioRxiv [Preprint]. 2022 Aug 2:2022.08.02.501850. doi: 10.1101/2022.08.02.501850. bioRxiv. 2022. Update in: J Proteome Res. 2023 Apr 7;22(4):1138-1147. doi: 10.1021/acs.jproteome.2c00475. PMID: 35982648 Free PMC article. Updated. Preprint.
-
A multiplexed microflow LC-MS/MS-PRM assay for serologic quantification of IgG N- and HPX O- glycoforms in liver fibrosis.Sci Rep. 2023 Jan 12;13(1):606. doi: 10.1038/s41598-023-27382-0. Sci Rep. 2023. PMID: 36635317 Free PMC article.
-
LC-MS3 quantification of O-glycopeptides in human serum.Electrophoresis. 2013 Aug;34(16):2342-9. doi: 10.1002/elps.201200658. Epub 2013 Jul 24. Electrophoresis. 2013. PMID: 23765987 Free PMC article.
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
-
Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides.Glycoconj J. 2016 Jun;33(3):261-72. doi: 10.1007/s10719-016-9649-3. Epub 2016 Jan 18. Glycoconj J. 2016. PMID: 26780731 Review.
Cited by
-
Serum N-Glycan Changes in Rats Chronically Exposed to Glyphosate-Based Herbicides.Biomolecules. 2024 Aug 28;14(9):1077. doi: 10.3390/biom14091077. Biomolecules. 2024. PMID: 39334844 Free PMC article.
-
Technical, preclinical, and clinical developments of Fc-glycan-specific antibody-drug conjugates.RSC Med Chem. 2024 Oct 18;16(1):50-62. doi: 10.1039/d4md00637b. eCollection 2025 Jan 23. RSC Med Chem. 2024. PMID: 39568595 Free PMC article. Review.
-
Simple Routes to Stable Isotope-Coded Native Glycans.Anal Chem. 2024 Jan 9;96(1):163-169. doi: 10.1021/acs.analchem.3c03446. Epub 2023 Dec 28. Anal Chem. 2024. PMID: 38153380 Free PMC article.
-
Targeted Glycoproteomics Analysis Using MRM/PRM Approaches.Methods Mol Biol. 2024;2762:231-250. doi: 10.1007/978-1-0716-3666-4_14. Methods Mol Biol. 2024. PMID: 38315369 Free PMC article.
-
Enhancing allergy diagnosis: mass spectrometry as a complementary technique to the basophil activation test.Front Allergy. 2025 May 30;6:1568670. doi: 10.3389/falgy.2025.1568670. eCollection 2025. Front Allergy. 2025. PMID: 40519814 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

