Epitranscriptomic regulation of cortical neurogenesis via Mettl8-dependent mitochondrial tRNA m3C modification
- PMID: 36764294
- PMCID: PMC10031801
- DOI: 10.1016/j.stem.2023.01.007
Epitranscriptomic regulation of cortical neurogenesis via Mettl8-dependent mitochondrial tRNA m3C modification
Abstract
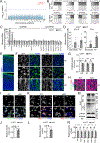
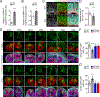
Increasing evidence implicates the critical roles of various epitranscriptomic RNA modifications in different biological processes. Methyltransferase METTL8 installs 3-methylcytosine (m3C) modification of mitochondrial tRNAs in vitro; however, its role in intact biological systems is unknown. Here, we show that Mettl8 is localized in mitochondria and installs m3C specifically on mitochondrial tRNAThr/Ser(UCN) in mouse embryonic cortical neural stem cells. At molecular and cellular levels, Mettl8 deletion in cortical neural stem cells leads to reduced mitochondrial protein translation and attenuated respiration activity. At the functional level, conditional Mettl8 deletion in mice results in impaired embryonic cortical neural stem cell maintenance in vivo, which can be rescued by pharmacologically enhancing mitochondrial functions. Similarly, METTL8 promotes mitochondrial protein expression and neural stem cell maintenance in human forebrain cortical organoids. Together, our study reveals a conserved epitranscriptomic mechanism of Mettl8 and mitochondrial tRNA m3C modification in maintaining embryonic cortical neural stem cells in mice and humans.
Keywords: Mettl8; epitranscriptomics; human forebrain organoids; m(3)C modification; mitochondria activity; mitochondrial tRNA; neural stem cells; neurogenesis.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests G.-l.M. is on the advisory board of Cell Stem Cell.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

