TET1 and TDG Suppress Inflammatory Response in Intestinal Tumorigenesis: Implications for Colorectal Tumors With the CpG Island Methylator Phenotype
- PMID: 36764492
- PMCID: PMC10586516
- DOI: 10.1053/j.gastro.2023.01.039
TET1 and TDG Suppress Inflammatory Response in Intestinal Tumorigenesis: Implications for Colorectal Tumors With the CpG Island Methylator Phenotype
Abstract
Background & aims: Aberrant DNA methylation is frequent in colorectal cancer (CRC), but underlying mechanisms and pathologic consequences are poorly understood.
Methods: We disrupted active DNA demethylation genes Tet1 and/or Tdg from ApcMin mice and characterized the methylome and transcriptome of colonic adenomas. Data were compared to human colonic adenocarcinomas (COAD) in The Cancer Genome Atlas.
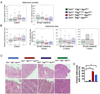
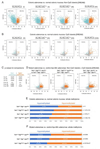
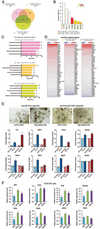
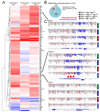
Results: There were increased numbers of small intestinal adenomas in ApcMin mice expressing the TdgN151A allele, whereas Tet1-deficient and Tet1/TdgN151A-double heterozygous ApcMin colonic adenomas were larger with features of erosion and invasion. We detected reduction in global DNA hypomethylation in colonic adenomas from Tet1- and Tdg-mutant ApcMin mice and hypermethylation of CpG islands in Tet1-mutant ApcMin adenomas. Up-regulation of inflammatory, immune, and interferon response genes was present in Tet1- and Tdg-mutant colonic adenomas compared to control ApcMin adenomas. This up-regulation was also seen in murine colonic organoids and human CRC lines infected with lentiviruses expressing TET1 or TDG short hairpin RNA. A 127-gene inflammatory signature separated colonic adenocarcinomas into 4 groups, closely aligned with their microsatellite or chromosomal instability and characterized by different levels of DNA methylation and DNMT1 expression that anticorrelated with TET1 expression. Tumors with the CpG island methylator phenotype (CIMP) had concerted high DNMT1/low TET1 expression. TET1 or TDG knockdown in CRC lines enhanced killing by natural killer cells.
Conclusions: Our findings reveal a novel epigenetic regulation, linked to the type of genomic instability, by which TET1/TDG-mediated DNA demethylation decreases methylation levels and inflammatory/interferon/immune responses. CIMP in CRC is triggered by an imbalance of methylating activities over demethylating activities. These mice represent a model of CIMP CRC.
Keywords: CpG Island Methylator Phenotype; DNA Demethylation; DNA Methylation; Inflammatory Response; Interferon Response.
Copyright © 2023 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest
J.R.W. has served or is serving as a consultant or advisor for Qsonica, Salarius Pharmaceuticals, Daiichi Sankyo, Inc., Vyne Therapeutics and Lily Asia Ventures. J.R.W also receives funding for research from Salarius Pharmaceuticals and Oryzon Genomics. No relevant conflicts of interest exist for the other authors.
Figures



References
-
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. - PubMed
-
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–767. - PubMed
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004;4:143–53. - PubMed
-
- Bellacosa A Genetic hits and mutation rate in colorectal tumorigenesis: versatility of Knudson’s theory and implications for cancer prevention. Genes Chromosomes Cancer 2003;38:382–388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

