Structure of VanS from vancomycin-resistant enterococci: A sensor kinase with weak ATP binding
- PMID: 36764524
- PMCID: PMC10017428
- DOI: 10.1016/j.jbc.2023.103001
Structure of VanS from vancomycin-resistant enterococci: A sensor kinase with weak ATP binding
Abstract
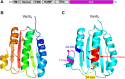
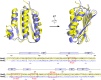
The VanRS two-component system regulates the resistance phenotype of vancomycin-resistant enterococci. VanS is a sensor histidine kinase that responds to the presence of vancomycin by autophosphorylating and subsequently transferring the phosphoryl group to the response regulator, VanR. The phosphotransfer activates VanR as a transcription factor, which initiates the expression of resistance genes. Structural information about VanS proteins has remained elusive, hindering the molecular-level understanding of their function. Here, we present X-ray crystal structures for the catalytic and ATP-binding (CA) domains of two VanS proteins, derived from vancomycin-resistant enterococci types A and C. Both proteins adopt the canonical Bergerat fold that has been observed for CA domains of other prokaryotic histidine kinases. We attempted to determine structures for the nucleotide-bound forms of both proteins; however, despite repeated efforts, these forms could not be crystallized, prompting us to measure the proteins' binding affinities for ATP. Unexpectedly, both CA domains displayed low affinities for the nucleotide, with KD values in the low millimolar range. Since these KD values are comparable to intracellular ATP concentrations, this weak substrate binding could reflect a way of regulating expression of the resistance phenotype.
Keywords: ATP binding; antibiotic resistance; histidine kinase; two-component system; vancomycin-resistant enterococci.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Levine D.P. Vancomycin: a history. Clin. Infect. Dis. 2006;42:S5–S12. - PubMed
-
- Joshi S., Shallal A., Zervos M. Vancomycin-resistant Enterococci: epidemiology, infection prevention, and control. Infect Dis. Clin. North Am. 2021;35:953–968. - PubMed
-
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. - PubMed
-
- Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect Dis. 2008;197:1079–1081. - PubMed
-
- Loll P.J., Axelsen P.H. The structural biology of molecular recognition by vancomycin. Annu. Rev. Biophys. Biomol. Struct. 2000;29:265–289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

