Bile salt hydrolase in non-enterotoxigenic Bacteroides potentiates colorectal cancer
- PMID: 36765047
- PMCID: PMC9918522
- DOI: 10.1038/s41467-023-36089-9
Bile salt hydrolase in non-enterotoxigenic Bacteroides potentiates colorectal cancer
Abstract
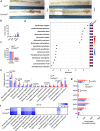
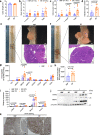
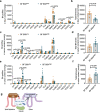
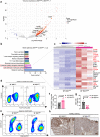
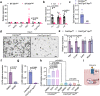
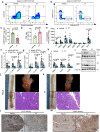
Bile salt hydrolase (BSH) in Bacteroides is considered a potential drug target for obesity-related metabolic diseases, but its involvement in colon tumorigenesis has not been explored. BSH-expressing Bacteroides is found at high abundance in the stools of colorectal cancer (CRC) patients with overweight and in the feces of a high-fat diet (HFD)-induced CRC mouse model. Colonization of B. fragilis 638R, a strain with low BSH activity, overexpressing a recombinant bsh gene from B. fragilis NCTC9343 strain, results in increased unconjugated bile acids in the colon and accelerated progression of CRC under HFD treatment. In the presence of high BSH activity, the resultant elevation of unconjugated deoxycholic acid and lithocholic acid activates the G-protein-coupled bile acid receptor, resulting in increased β-catenin-regulated chemokine (C-C motif) ligand 28 (CCL28) expression in colon tumors. Activation of the β-catenin/CCL28 axis leads to elevated intra-tumoral immunosuppressive CD25+FOXP3+ Treg cells. Blockade of the β-catenin/CCL28 axis releases the immunosuppression to enhance the intra-tumoral anti-tumor response, which decreases CRC progression under HFD treatment. Pharmacological inhibition of BSH reduces HFD-accelerated CRC progression, coincident with suppression of the β-catenin/CCL28 pathway. These findings provide insights into the pro-carcinogenetic role of Bacteroides in obesity-related CRC progression and characterize BSH as a potential target for CRC prevention and treatment.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Siegel RL, et al. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020;70:145–164. - PubMed
-
- Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 2001;1:55–67. - PubMed
-
- Romano G, Chagani S, Kwong LN. The path to metastatic mouse models of colorectal cancer. Oncogene. 2018;37:2481–2489. - PubMed
-
- Garrett WS. The gut microbiota and colon cancer. Science. 2019;364:1133–1135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

