Differential expression patterns of long noncoding RNAs in a pleiomorphic diatom and relation to hyposalinity
- PMID: 36765079
- PMCID: PMC9918465
- DOI: 10.1038/s41598-023-29489-w
Differential expression patterns of long noncoding RNAs in a pleiomorphic diatom and relation to hyposalinity
Abstract
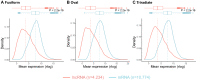
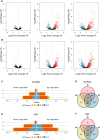
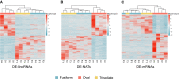
Long non-coding (lnc)RNAs have been shown to have central roles in stress responses, cell identity and developmental processes in multicellular organisms as well as in unicellular fungi. Previous works have shown the occurrence of lncRNAs in diatoms, namely in Phaeodactylum tricornutum, many of which being expressed under specific stress conditions. Interestingly, P. tricornutum is the only known diatom that has a demonstrated morphological plasticity, occurring in three distinct morphotypes: fusiform, triradiate and oval. Although the morphotypes are interchangeable, the fusiform is the dominant one while both the triradiate and the oval forms are less common, the latter often being associated with stress conditions such as low salinity and solid culture media, amongst others. Nonetheless, the molecular basis underpinning morphotype identity in P. tricornutum remains elusive. Using twelve previously published transcriptomic datasets originating from the three morphotypes of P. tricornutum, we sought to investigate the expression patterns of lncRNAs (lincRNAs and NATs) in these distinct morphotypes, using pairwise comparisons, in order to explore the putative involvement of these noncoding molecules in morphotype identity. We found that differentially expressed lncRNAs cluster according to morphotype, indicating that lncRNAs are not randomly expressed, but rather seem to provide a specific (noncoding) transcriptomic signature of the morphotype. We also present evidence to suggest that the major differences in DE genes (both noncoding and coding) between the stress related oval morphotype and the most common fusiform morphotype could be due, to a large extent, to the hyposaline culture conditions rather than to the morphotype itself. However, several lncRNAs associated to each one of the three morphotypes were identified, which could have a potential role in morphotype (or cell) identity in P. tricornutum, similar to what has been found in both animals and plant development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Sims PA, Mann DG, Medlin LK. Evolution of the diatoms: Insights from fossil, biological and molecular data. Phycologia. 2006;45:361–402. doi: 10.2216/05-22.1. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

