Long-term platinum-based drug accumulation in cancer-associated fibroblasts promotes colorectal cancer progression and resistance to therapy
- PMID: 36765091
- PMCID: PMC9918738
- DOI: 10.1038/s41467-023-36334-1
Long-term platinum-based drug accumulation in cancer-associated fibroblasts promotes colorectal cancer progression and resistance to therapy
Abstract
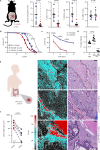
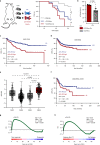
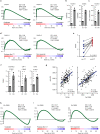
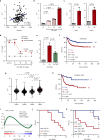
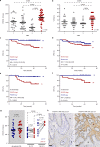
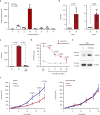
A substantial proportion of cancer patients do not benefit from platinum-based chemotherapy (CT) due to the emergence of drug resistance. Here, we apply elemental imaging to the mapping of CT biodistribution after therapy in residual colorectal cancer and achieve a comprehensive analysis of the genetic program induced by oxaliplatin-based CT in the tumor microenvironment. We show that oxaliplatin is largely retained by cancer-associated fibroblasts (CAFs) long time after the treatment ceased. We determine that CT accumulation in CAFs intensifies TGF-beta activity, leading to the production of multiple factors enhancing cancer aggressiveness. We establish periostin as a stromal marker of chemotherapeutic activity intrinsically upregulated in consensus molecular subtype 4 (CMS4) tumors and highly expressed before and/or after treatment in patients unresponsive to therapy. Collectively, our study underscores the ability of CT-retaining CAFs to support cancer progression and resistance to treatment.
© 2023. The Author(s).
Conflict of interest statement
A.Ce. declares institutional research funding from Genentech, Merck Serono, Bristol Myers Squibb, Merck Sharp & Dohme, Roche, Beigene, Bayer, Servier, Lilly, Novartis, Takeda, Astellas, Takeda and Fibrogen; and advisory board or speaker fees from Amgen, Merck Serono, Roche, Bayer, Servier and Pierre Fabre in the last 5 years. J.T. reports personal financial interest in form of scientific consultancy role for Array Biopharma, AstraZeneca, Bayer, Boehringer Ingelheim, Chugai, Daiichi Sankyo, F. Hoffmann-La Roche Ltd, Genentech Inc, HalioDX SAS, Hutchison MediPharma International, Ikena Oncology, Inspirna Inc, IQVIA, Lilly, Menarini, Merck Serono, Merus, MSD, Mirati, Neophore, Novartis, Ona Therapeutics, Orion Biotechnology, Peptomyc, Pfizer, Pierre Fabre, Samsung Bioepis, Sanofi, Scandion Oncology, Scorpion Therapeutics, Seattle Genetics, Servier, Sotio Biotech, Taiho, Tessa Therapeutics and TheraMyc. Stocks: Oniria Therapeutics and also educational collaboration with Imedex/HMP, Medscape Education, MJH Life Sciences, PeerView Institute for Medical Education and Physicians Education Resource (PER). N.T. declares the following: Advisory Role: Merck Serono, Guardant Health, Speaking: Amgen, Servier, Pfizer, Merck Serono, ESMO, SEOM. C.M. reports personal financial interest in form of scientific consultancy role for Amgen, Biocartis, F. Hoffmann-La Roche Ltd, Genentech Inc, Merck Serono, Pfizer, Pierre Fabre, Sanofi, also educational collaboration with Amgen, Guardant Health, Merck Serono. The remaining authors declare no other competing interests.
Figures

References
-
- Rottenberg S, Disler C, Perego P. The rediscovery of platinum-based cancer therapy. Nat. Rev. Cancer. 2020;211:37–50. - PubMed
-
- Schmoll HJ, et al. Effect of adjuvant capecitabine or fluorouracil, with or without oxaliplatin, on survival outcomes in stage III colon cancer and the effect of oxaliplatin on post-relapse survival: a pooled analysis of individual patient data from four randomised controlled trials. Lancet Oncol. 2014;15:1481–1492. - PMC - PubMed
-
- Cunningham D, et al. Colorectal cancer. Lancet. 2010;375:1030–1047. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

