Region Segmentation of Whole-Slide Images for Analyzing Histological Differentiation of Prostate Adenocarcinoma Using Ensemble EfficientNetB2 U-Net with Transfer Learning Mechanism
- PMID: 36765719
- PMCID: PMC9913745
- DOI: 10.3390/cancers15030762
Region Segmentation of Whole-Slide Images for Analyzing Histological Differentiation of Prostate Adenocarcinoma Using Ensemble EfficientNetB2 U-Net with Transfer Learning Mechanism
Abstract
Recent advances in computer-aided detection via deep learning (DL) now allow for prostate cancer to be detected automatically and recognized with extremely high accuracy, much like other medical diagnoses and prognoses. However, researchers are still limited by the Gleason scoring system. The histopathological analysis involved in assigning the appropriate score is a rigorous, time-consuming manual process that is constrained by the quality of the material and the pathologist's level of expertise. In this research, we implemented a DL model using transfer learning on a set of histopathological images to segment cancerous and noncancerous areas in whole-slide images (WSIs). In this approach, the proposed Ensemble U-net model was applied for the segmentation of stroma, cancerous, and benign areas. The WSI dataset of prostate cancer was collected from the Kaggle repository, which is publicly available online. A total of 1000 WSIs were used for region segmentation. From this, 8100 patch images were used for training, and 900 for testing. The proposed model demonstrated an average dice coefficient (DC), intersection over union (IoU), and Hausdorff distance of 0.891, 0.811, and 15.9, respectively, on the test set, with corresponding masks of patch images. The manipulation of the proposed segmentation model improves the ability of the pathologist to predict disease outcomes, thus enhancing treatment efficacy by isolating the cancerous regions in WSIs.
Keywords: U-Net; deep learning; histological; prostate adenocarcinoma; segmentation; transfer learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



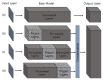
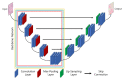

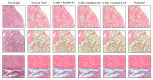
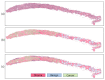
References
-
- Gnanapragasam V.J., Bratt O., Muir K., Lee L.S., Huang H.H., Stattin P., Lophatananon A. The Cambridge Prognostic Groups for Improved Prediction of Disease Mortality at Diagnosis in Primary Non-Metastatic Prostate Cancer: A Validation Study. BMC Med. 2018;16:31. doi: 10.1186/s12916-018-1019-5. - DOI - PMC - PubMed
-
- Daskivich T.J., Fan K.-H., Koyama T., Albertsen P.C., Goodman M., Hamilton A.S., Hoffman R.M., Stanford J.L., Stroup A.M., Litwin M.S., et al. Prediction of Long-Term Other-Cause Mortality in Men with Early-Stage Prostate Cancer: Results from the Prostate Cancer Outcomes Study. Urology. 2015;85:92–100. doi: 10.1016/j.urology.2014.07.003. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

