Aurora B SUMOylation Is Restricted to Centromeres in Early Mitosis and Requires RANBP2
- PMID: 36766713
- PMCID: PMC9913629
- DOI: 10.3390/cells12030372
Aurora B SUMOylation Is Restricted to Centromeres in Early Mitosis and Requires RANBP2
Abstract
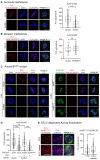
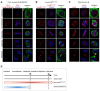
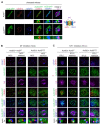
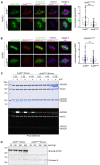
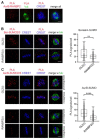
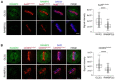
Conjugation with the small ubiquitin-like modifier (SUMO) modulates protein interactions and localisation. The kinase Aurora B, a key regulator of mitosis, was previously identified as a SUMOylation target in vitro and in assays with overexpressed components. However, where and when this modification genuinely occurs in human cells was not ascertained. Here, we have developed intramolecular Proximity Ligation Assays (PLA) to visualise SUMO-conjugated Aurora B in human cells in situ. We visualised Aurora B-SUMO products at centromeres in prometaphase and metaphase, which declined from anaphase onwards and became virtually undetectable at cytokinesis. In the mitotic window in which Aurora B/SUMO products are abundant, Aurora B co-localised and interacted with NUP358/RANBP2, a nucleoporin with SUMO ligase and SUMO-stabilising activity. Indeed, in addition to the requirement for the previously identified PIAS3 SUMO ligase, we found that NUP358/RANBP2 is also implicated in Aurora B-SUMO PLA product formation and centromere localisation. In summary, SUMOylation marks a distinctive window of Aurora B functions at centromeres in prometaphase and metaphase while being dispensable for functions exerted in cytokinesis, and RANBP2 contributes to this control, adding a novel layer to modulation of Aurora B functions during mitosis.
Keywords: Aurora B; RANBP2; SUMOylation; in situ proximity ligation assay (isPLA); mitosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Wotton D., Pemberton L.F., Merrill-Schools J. SUMO and Chromatin Remodeling. Adv. Exp. Med. Biol. 2017;963:35–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

