S-Palmitoylation of Synaptic Proteins in Neuronal Plasticity in Normal and Pathological Brains
- PMID: 36766729
- PMCID: PMC9913408
- DOI: 10.3390/cells12030387
S-Palmitoylation of Synaptic Proteins in Neuronal Plasticity in Normal and Pathological Brains
Abstract
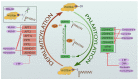
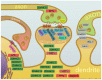
Protein lipidation is a common post-translational modification of proteins that plays an important role in human physiology and pathology. One form of protein lipidation, S-palmitoylation, involves the addition of a 16-carbon fatty acid (palmitate) onto proteins. This reversible modification may affect the regulation of protein trafficking and stability in membranes. From multiple recent experimental studies, a picture emerges whereby protein S-palmitoylation is a ubiquitous yet discrete molecular switch enabling the expansion of protein functions and subcellular localization in minutes to hours. Neural tissue is particularly rich in proteins that are regulated by S-palmitoylation. A surge of novel methods of detection of protein lipidation at high resolution allowed us to get better insights into the roles of protein palmitoylation in brain physiology and pathophysiology. In this review, we specifically discuss experimental work devoted to understanding the impact of protein palmitoylation on functional changes in the excitatory and inhibitory synapses associated with neuronal activity and neuronal plasticity. The accumulated evidence also implies a crucial role of S-palmitoylation in learning and memory, and brain disorders associated with impaired cognitive functions.
Keywords: S-palmitoylation; brain disorders; learning and memory; synaptic plasticity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

