Interaction of Tryptophan- and Arginine-Rich Antimicrobial Peptide with E. coli Outer Membrane-A Molecular Simulation Approach
- PMID: 36768325
- PMCID: PMC9916935
- DOI: 10.3390/ijms24032005
Interaction of Tryptophan- and Arginine-Rich Antimicrobial Peptide with E. coli Outer Membrane-A Molecular Simulation Approach
Abstract
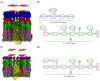
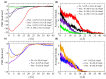
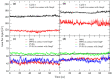
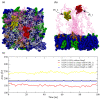
A short antimicrobial peptide (AMP), rich in tryptophan and arginine (P6-HRWWRWWRR-NH2), was used in molecular dynamics (MD) simulations to investigate the interaction between AMPs and lipopolysaccharides (LPS) from two E. coli outer membrane (OM) membrane models. The OM of Gram-negative bacteria is an asymmetric bilayer, with the outer layer consisting exclusively of lipopolysaccharide molecules and the lower leaflet made up of phospholipids. The mechanisms by which short AMPs permeate the OM of Gram-negative bacteria are not well understood at the moment. For this study, two types of E. coli OM membrane models were built with (i) smooth LPS composed of lipid A, K12 core and O21 O-antigen, and (ii) rough type LPS composed of lipid A and R1 core. An OmpF monomer from E. coli was embedded in both membrane models. MD trajectories revealed that AMP insertion in the LPS layer was facilitated by the OmpF-created gap and allowed AMPs to form hydrogen bonds with the phosphate groups of inner core oligosaccharides. OM proteins such as OmpF may be essential for the permeation of short AMPs such as P6 by exposing the LPS binding site or even by direct translocation of AMPs across the OM.
Keywords: E. coli; MD; PMF; antimicrobial-peptide; lipopolysaccharide; outer membrane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

