The Repertoire of RNA Modifications Orchestrates a Plethora of Cellular Responses
- PMID: 36768716
- PMCID: PMC9916637
- DOI: 10.3390/ijms24032387
The Repertoire of RNA Modifications Orchestrates a Plethora of Cellular Responses
Abstract
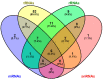
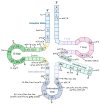
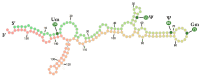
Although a plethora of DNA modifications have been extensively investigated in the last decade, recent breakthroughs in molecular biology, including high throughput sequencing techniques, have enabled the identification of post-transcriptional marks that decorate RNAs; hence, epitranscriptomics has arisen. This recent scientific field aims to decode the regulatory layer of the transcriptome and set the ground for the detection of modifications in ribose nucleotides. Until now, more than 170 RNA modifications have been reported in diverse types of RNA that contribute to various biological processes, such as RNA biogenesis, stability, and transcriptional and translational accuracy. However, dysfunctions in the RNA-modifying enzymes that regulate their dynamic level can lead to human diseases and cancer. The present review aims to highlight the epitranscriptomic landscape in human RNAs and match the catalytic proteins with the deposition or deletion of a specific mark. In the current review, the most abundant RNA modifications, such as N6-methyladenosine (m6A), N5-methylcytosine (m5C), pseudouridine (Ψ) and inosine (I), are thoroughly described, their functional and regulatory roles are discussed and their contributions to cellular homeostasis are stated. Ultimately, the involvement of the RNA modifications and their writers, erasers, and readers in human diseases and cancer is also discussed.
Keywords: alternative splicing; erasers; m5C; m6A; mRNA stability; post-transcriptional modifications; readers; translation efficiency; writers; Ψ.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Deciphering the epitranscriptome: A green perspective.J Integr Plant Biol. 2016 Oct;58(10):822-835. doi: 10.1111/jipb.12483. Epub 2016 Jun 20. J Integr Plant Biol. 2016. PMID: 27172004 Free PMC article. Review.
-
The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities.Mol Cancer. 2021 Jan 18;20(1):18. doi: 10.1186/s12943-020-01263-w. Mol Cancer. 2021. PMID: 33461542 Free PMC article. Review.
-
The Dark Side of the Epitranscriptome: Chemical Modifications in Long Non-Coding RNAs.Int J Mol Sci. 2017 Nov 10;18(11):2387. doi: 10.3390/ijms18112387. Int J Mol Sci. 2017. PMID: 29125541 Free PMC article. Review.
-
Deciphering the Epitranscriptomic Signatures in Cell Fate Determination and Development.Stem Cell Rev Rep. 2019 Aug;15(4):474-496. doi: 10.1007/s12015-019-09894-3. Stem Cell Rev Rep. 2019. PMID: 31123982 Review.
-
Modifications of RNA in cancer: a comprehensive review.Mol Biol Rep. 2025 Mar 17;52(1):321. doi: 10.1007/s11033-025-10419-0. Mol Biol Rep. 2025. PMID: 40095076 Review.
Cited by
-
RNA modifications and their role in gene expression.Front Mol Biosci. 2025 Apr 25;12:1537861. doi: 10.3389/fmolb.2025.1537861. eCollection 2025. Front Mol Biosci. 2025. PMID: 40351534 Free PMC article. Review.
-
Epitranscriptomic alterations induced by environmental toxins: implications for RNA modifications and disease.Genes Environ. 2025 Aug 4;47(1):14. doi: 10.1186/s41021-025-00337-9. Genes Environ. 2025. PMID: 40760453 Free PMC article. Review.
-
Urine metabolomics signature reveals novel determinants of adrenal suppression in children taking inhaled corticosteroids to control asthma symptoms.Immun Inflamm Dis. 2024 Jul;12(7):e1315. doi: 10.1002/iid3.1315. Immun Inflamm Dis. 2024. PMID: 39031511 Free PMC article.
-
Deaminase-Driven Reverse Transcription Mutagenesis in Oncogenesis: Critical Analysis of Transcriptional Strand Asymmetries of Single Base Substitution Signatures.Int J Mol Sci. 2025 Jan 24;26(3):989. doi: 10.3390/ijms26030989. Int J Mol Sci. 2025. PMID: 39940758 Free PMC article. Review.
-
RNA Through Time: From the Origin of Life to Therapeutic Frontiers in Transcriptomics and Epitranscriptional Medicine.Int J Mol Sci. 2025 May 22;26(11):4964. doi: 10.3390/ijms26114964. Int J Mol Sci. 2025. PMID: 40507776 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

