Big Data in Gastroenterology Research
- PMID: 36768780
- PMCID: PMC9916510
- DOI: 10.3390/ijms24032458
Big Data in Gastroenterology Research
Abstract
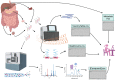
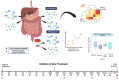
Studying individual data types in isolation provides only limited and incomplete answers to complex biological questions and particularly falls short in revealing sufficient mechanistic and kinetic details. In contrast, multi-omics approaches to studying health and disease permit the generation and integration of multiple data types on a much larger scale, offering a comprehensive picture of biological and disease processes. Gastroenterology and hepatobiliary research are particularly well-suited to such analyses, given the unique position of the luminal gastrointestinal (GI) tract at the nexus between the gut (mucosa and luminal contents), brain, immune and endocrine systems, and GI microbiome. The generation of 'big data' from multi-omic, multi-site studies can enhance investigations into the connections between these organ systems and organisms and more broadly and accurately appraise the effects of dietary, pharmacological, and other therapeutic interventions. In this review, we describe a variety of useful omics approaches and how they can be integrated to provide a holistic depiction of the human and microbial genetic and proteomic changes underlying physiological and pathophysiological phenomena. We highlight the potential pitfalls and alternatives to help avoid the common errors in study design, execution, and analysis. We focus on the application, integration, and analysis of big data in gastroenterology and hepatobiliary research.
Keywords: epigenomics; genomics; gut microbiome; medical informatics; metabolomics; proteomics; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

