The Journey of Mitochondrial Protein Import and the Roadmap to Follow
- PMID: 36768800
- PMCID: PMC9916854
- DOI: 10.3390/ijms24032479
The Journey of Mitochondrial Protein Import and the Roadmap to Follow
Abstract
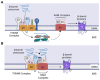
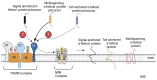
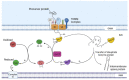
Mitochondria are double membrane-bound organelles that play critical functions in cells including metabolism, energy production, regulation of intrinsic apoptosis, and maintenance of calcium homeostasis. Mitochondria are fascinatingly equipped with their own genome and machinery for transcribing and translating 13 essential proteins of the oxidative phosphorylation system (OXPHOS). The rest of the proteins (99%) that function in mitochondria in the various pathways described above are nuclear-transcribed and synthesized as precursors in the cytosol. These proteins are imported into the mitochondria by the unique mitochondrial protein import system that consists of seven machineries. Proper functioning of the mitochondrial protein import system is crucial for optimal mitochondrial deliverables, as well as mitochondrial and cellular homeostasis. Impaired mitochondrial protein import leads to proteotoxic stress in both mitochondria and cytosol, inducing mitochondrial unfolded protein response (UPRmt). Altered UPRmt is associated with the development of various disease conditions including neurodegenerative and cardiovascular diseases, as well as cancer. This review sheds light on the molecular mechanisms underlying the import of nuclear-encoded mitochondrial proteins, the consequences of defective mitochondrial protein import, and the pathological conditions that arise due to altered UPRmt.
Keywords: diseases; mitochondria; mitochondrial protein import machineries; mitochondrial unfolded protein response; proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wang Z., Ying Z., Bosy-Westphal A., Zhang J., Schautz B., Later W., Heymsfield S.B., Müller M.J. Specific metabolic rates of major organs and tissues across adulthood: Evaluation by mechanistic model of resting energy expenditure. Am. J. Clin. Nutr. 2010;92:1369–1377. doi: 10.3945/ajcn.2010.29885. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

