Dysregulation of Gene Expression of Key Signaling Mediators in PBMCs from People with Type 2 Diabetes Mellitus
- PMID: 36769056
- PMCID: PMC9916932
- DOI: 10.3390/ijms24032732
Dysregulation of Gene Expression of Key Signaling Mediators in PBMCs from People with Type 2 Diabetes Mellitus
Abstract
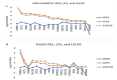
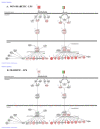
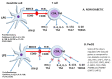
Diabetes is currently the fifth leading cause of death by disease in the USA. The underlying mechanisms for type 2 Diabetes Mellitus (DM2) and the enhanced susceptibility of such patients to inflammatory disorders and infections remain to be fully defined. We have recently shown that peripheral blood mononuclear cells (PBMCs) from non-diabetic people upregulate expression of inflammatory genes in response to proteasome modulators, such as bacterial lipopolysaccharide (LPS) and soybean lectin (LEC); in contrast, resveratrol (RES) downregulates this response. We hypothesized that LPS and LEC will also elicit a similar upregulation of gene expression of key signaling mediators in (PBMCs) from people with type 2 diabetes (PwD2, with chronic inflammation) ex vivo. Unexpectedly, using next generation sequencing (NGS), we show for the first time, that PBMCs from PwD2 failed to elicit a robust LPS- and LEC-induced gene expression of proteasome subunit LMP7 (PSMB8) and mediators of T cell signaling that were observed in non-diabetic controls. These repressed genes included: PSMB8, PSMB9, interferon-γ, interferon-λ, signal-transducer-and-activator-of-transcription-1 (STAT1), human leukocyte antigen (HLA DQB1, HLA DQA1) molecules, interleukin 12A, tumor necrosis factor-α, transporter associated with antigen processing 1 (TAP1), and several others, which showed a markedly weak upregulation with toxins in PBMCs from PwD2, as compared to those from non-diabetics. Resveratrol (proteasome inhibitor) further downregulated the gene expression of these inflammatory mediators in PBMCs from PwD2. These results might explain why PwD2 may be susceptible to infectious disease. LPS and toxins may be leading to inflammation, insulin resistance, and thus, metabolic changes in the host cells.
Keywords: IFN-γ; LPS; NGS; NO; cytokines; lectins; resveratrol; signal transduction; type 2 diabetes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Jesenak M., Brndiarova M., Urbancikova I., Rennerova Z., Vojtkova J., Bobcakova A., Ostro R., Banovcin P. Immune parameters, and COVID-19 infection-associations with clinical severity and diseases prognosis. Front. Cell. Infect. Microbiology. 2020;10:364. doi: 10.3389/fcimb.2020.00364. - DOI - PMC - PubMed
-
- Siddiqui A.A., Siddiqui S.A., Ahmad S., Siddiqui S., Ahsan I., Sahu K. Diabetes: Mechanism, pathophysiology, and management-A review. Int. J. Drug Dev. Res. 2013;5:1–23.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

