Rapid One-Step Capturing of Native, Cell-Free Synthesized and Membrane-Embedded GLP-1R
- PMID: 36769142
- PMCID: PMC9917595
- DOI: 10.3390/ijms24032808
Rapid One-Step Capturing of Native, Cell-Free Synthesized and Membrane-Embedded GLP-1R
Abstract
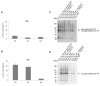
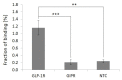
G protein-coupled receptors (GPCRs) are of outstanding pharmacological interest as they are abundant in cell membranes where they perform diverse functions that are closely related to the vitality of cells. The analysis of GPCRs in natural membranes is laborious, as established methods are almost exclusively cell culture-based and only a few methods for immobilization in a natural membrane outside the cell are known. Within this study, we present a one-step, fast and robust immobilization strategy of the GPCR glucagon-like peptide 1 receptor (GLP-1R). GLP-1R was synthesized in eukaryotic lysates harboring endogenous endoplasmic reticulum-derived microsomes enabling the embedment of GLP-1R in a natural membrane. Interestingly, we found that these microsomes spontaneously adsorbed to magnetic Neutravidin beads thus providing immobilized membrane protein preparations which required no additional manipulation of the target receptor or its supporting membrane. The accessibility of the extracellular domain of membrane-embedded and bead-immobilized GLP-1R was demonstrated by bead-based enzyme-linked immunosorbent assay (ELISA) using GLP-1R-specific monoclonal antibodies. In addition, ligand binding of immobilized GLP-1R was verified in a radioligand binding assay. In summary, we present an easy and straightforward synthesis and immobilization methodology of an active GPCR which can be beneficial for studying membrane proteins in general.
Keywords: CFPS; G protein-coupled receptors; GLP-1R; Sf21 cell lysate; cell-free expression; cell-free protein synthesis; immobilization to magnetic beads; membrane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

