Emerging Role of SMILE in Liver Metabolism
- PMID: 36769229
- PMCID: PMC9917820
- DOI: 10.3390/ijms24032907
Emerging Role of SMILE in Liver Metabolism
Abstract
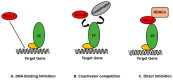
Small heterodimer partner-interacting leucine zipper (SMILE) is a member of the CREB/ATF family of basic leucine zipper (bZIP) transcription factors. SMILE has two isoforms, a small and long isoform, resulting from alternative usage of the initiation codon. Interestingly, although SMILE can homodimerize similar to other bZIP proteins, it cannot bind to DNA. As a result, SMILE acts as a co-repressor in nuclear receptor signaling and other transcription factors through its DNA binding inhibition, coactivator competition, and direct repression, thereby regulating the expression of target genes. Therefore, the knockdown of SMILE increases the transactivation of transcription factors. Recent findings suggest that SMILE is an important regulator of metabolic signals and pathways by causing changes in glucose, lipid, and iron metabolism in the liver. The regulation of SMILE plays an important role in pathological conditions such as hepatitis, diabetes, fatty liver disease, and controlling the energy metabolism in the liver. This review focuses on the role of SMILE and its repressive actions on the transcriptional activity of nuclear receptors and bZIP transcription factors and its effects on liver metabolism. Understanding the importance of SMILE in liver metabolism and signaling pathways paves the way to utilize SMILE as a target in treating liver diseases.
Keywords: CREBZF; SMILE; bZIP proteins; liver; metabolic pathways; nuclear receptors; transcription factors.
Conflict of interest statement
The authors have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

