Hepatic Glucose Metabolism Disorder Induced by Adipose Tissue-Derived miR-548ag via DPP4 Upregulation
- PMID: 36769291
- PMCID: PMC9917501
- DOI: 10.3390/ijms24032964
Hepatic Glucose Metabolism Disorder Induced by Adipose Tissue-Derived miR-548ag via DPP4 Upregulation
Abstract
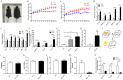
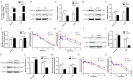
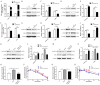
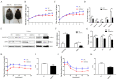
The present study aimed to explore the molecular mechanism underlying the regulation of glucose metabolism by miR-548ag. For the first time, we found that miR-548ag expression was elevated in the abdominal adipose tissue and serum of subjects with obesity and type 2 diabetes mellitus (T2DM). The conditional knockout of adipose tissue Dicer notably reduced the expression and content of miR-548ag in mouse adipose tissue, serum, and liver tissue. The combined use of RNAseq, an miRNA target gene prediction software, and the dual luciferase reporter assay confirmed that miR-548ag exerts a targeted regulatory effect on DNMT3B and DPP4. miR-548ag and DPP4 expression was increased in the adipose tissue, serum, and liver tissue of diet-induced obese mice, while DNMT3B expression was decreased. It was subsequently confirmed both in vitro and in vivo that adipose tissue-derived miR-548ag impaired glucose tolerance and insulin sensitivity by inhibiting DNMT3B and upregulating DPP4. Moreover, miR-548ag inhibitors significantly improved the adverse metabolic phenotype in both obese mice and db/db mice. These results revealed that the expression of the adipose tissue-derived miR-548ag increased in obese subjects, and that this could upregulate the expression of DPP4 by targeting DNMT3B, ultimately leading to glucose metabolism disorder. Therefore, miR-548ag could be utilized as a potential target in the treatment of T2DM.
Keywords: DNMT3B; DPP4; T2DM; miR-548ag; obesity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Exosome-derived miR-548ag drives hepatic lipid accumulation via upregulating FASN through inhibition of DNMT3B.J Lipid Res. 2025 Jun;66(6):100818. doi: 10.1016/j.jlr.2025.100818. Epub 2025 May 6. J Lipid Res. 2025. PMID: 40339699 Free PMC article.
-
miR-548ag promotes DPP4 expression in hepatocytes through activation of TLR(7/8)/NF-κB pathway.Int J Obes (Lond). 2024 Jul;48(7):941-953. doi: 10.1038/s41366-024-01504-8. Epub 2024 Feb 29. Int J Obes (Lond). 2024. PMID: 38424257 Free PMC article.
-
DPP4 deletion in adipose tissue improves hepatic insulin sensitivity in diet-induced obesity.Am J Physiol Endocrinol Metab. 2020 May 1;318(5):E590-E599. doi: 10.1152/ajpendo.00323.2019. Epub 2019 Dec 31. Am J Physiol Endocrinol Metab. 2020. PMID: 31891536
-
Dipeptidyl peptidase inhibitor therapy in type 2 diabetes: Control of the incretin axis and regulation of postprandial glucose and lipid metabolism.Peptides. 2018 Feb;100:158-164. doi: 10.1016/j.peptides.2017.11.023. Peptides. 2018. PMID: 29412815 Review.
-
The Role of microRNA-22 in Metabolism.Int J Mol Sci. 2025 Jan 17;26(2):782. doi: 10.3390/ijms26020782. Int J Mol Sci. 2025. PMID: 39859495 Free PMC article. Review.
Cited by
-
Epigenetic modifications in obesity-associated diseases.MedComm (2020). 2024 Feb 24;5(2):e496. doi: 10.1002/mco2.496. eCollection 2024 Feb. MedComm (2020). 2024. PMID: 38405061 Free PMC article. Review.
-
Exosome-derived miR-548ag drives hepatic lipid accumulation via upregulating FASN through inhibition of DNMT3B.J Lipid Res. 2025 Jun;66(6):100818. doi: 10.1016/j.jlr.2025.100818. Epub 2025 May 6. J Lipid Res. 2025. PMID: 40339699 Free PMC article.
-
miR-548ag promotes DPP4 expression in hepatocytes through activation of TLR(7/8)/NF-κB pathway.Int J Obes (Lond). 2024 Jul;48(7):941-953. doi: 10.1038/s41366-024-01504-8. Epub 2024 Feb 29. Int J Obes (Lond). 2024. PMID: 38424257 Free PMC article.
-
Characterizing Circulating microRNA Signatures of Type 2 Diabetes Subtypes.Int J Mol Sci. 2025 Jan 14;26(2):637. doi: 10.3390/ijms26020637. Int J Mol Sci. 2025. PMID: 39859351 Free PMC article.
-
Adipose Tissue as a Major Launch Spot for Circulating Extracellular Vesicle-Carried MicroRNAs Coordinating Tissue and Systemic Metabolism.Int J Mol Sci. 2024 Dec 17;25(24):13488. doi: 10.3390/ijms252413488. Int J Mol Sci. 2024. PMID: 39769251 Free PMC article. Review.
References
-
- Frigolet M.E., Gutiérrez-Aguilar R. The colors of adipose tissue. Los colores del tejido adiposo. Gac. Médica México. 2020;156:142–149. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

