HERV-W ENV Induces Innate Immune Activation and Neuronal Apoptosis via linc01930/cGAS Axis in Recent-Onset Schizophrenia
- PMID: 36769337
- PMCID: PMC9917391
- DOI: 10.3390/ijms24033000
HERV-W ENV Induces Innate Immune Activation and Neuronal Apoptosis via linc01930/cGAS Axis in Recent-Onset Schizophrenia
Abstract
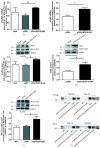
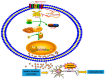
Schizophrenia is a severe neuropsychiatric disorder affecting about 1% of individuals worldwide. Increased innate immune activation and neuronal apoptosis are common findings in schizophrenia. Interferon beta (IFN-β), an essential cytokine in promoting and regulating innate immune responses, causes neuronal apoptosis in vitro. However, the precise pathogenesis of schizophrenia is unknown. Recent studies indicate that a domesticated endogenous retroviral envelope glycoprotein of the W family (HERV-W ENV, also called ERVWE1 or syncytin 1), derived from the endogenous retrovirus group W member 1 (ERVWE1) locus on chromosome 7q21.2, has a high level in schizophrenia. Here, we found an increased serum IFN-β level in schizophrenia and showed a positive correlation with HERV-W ENV. In addition, serum long intergenic non-protein coding RNA 1930 (linc01930), decreased in schizophrenia, was negatively correlated with HERV-W ENV and IFN-β. In vitro experiments showed that linc01930, mainly in the nucleus and with noncoding functions, was repressed by HERV-W ENV through promoter activity suppression. Further studies indicated that HERV-W ENV increased IFN-β expression and neuronal apoptosis by restraining the expression of linc01930. Furthermore, HERV-W ENV enhanced cyclic GMP-AMP synthase (cGAS) and stimulator of interferon genes protein (STING) expression and interferon regulatory factor 3 (IRF3) phosphorylation in neuronal cells. Notably, cGAS interacted with HERV-W ENV and triggered IFN-β expression and neuronal apoptosis caused by HERV-W ENV. Moreover, Linc01930 participated in the increased neuronal apoptosis and expression level of cGAS and IFN-β induced by HERV-W ENV. To summarize, our results suggested that linc01930 and IFN-β might be novel potential blood-based biomarkers in schizophrenia. The totality of these results also showed that HERV-W ENV facilitated antiviral innate immune response, resulting in neuronal apoptosis through the linc01930/cGAS/STING pathway in schizophrenia. Due to its monoclonal antibody GNbAC1 application in clinical trials, we considered HERV-W ENV might be a reliable therapeutic choice for schizophrenia.
Keywords: HERV-W ENV; IFN-β; apoptosis; cGAS; linc01930; schizophrenia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures












References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

