In Silico Targeting of Fascin Protein for Cancer Therapy: Benchmarking, Virtual Screening and Molecular Dynamics Approaches
- PMID: 36770963
- PMCID: PMC9921211
- DOI: 10.3390/molecules28031296
In Silico Targeting of Fascin Protein for Cancer Therapy: Benchmarking, Virtual Screening and Molecular Dynamics Approaches
Abstract
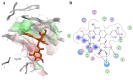
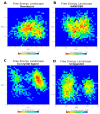
Fascin is an actin-bundling protein overexpressed in various invasive metastatic carcinomas through promoting cell migration and invasion. Therefore, blocking Fascin binding sites is considered a vital target for antimetastatic drugs. This inspired us to find new Fascin binding site blockers. First, we built an active compound set by collecting reported small molecules binding to Fascin's binding site 2. Consequently, a high-quality decoys set was generated employing DEKOIS 2.0 protocol to be applied in conducting the benchmarking analysis against the selected Fascin structures. Four docking programs, MOE, AutoDock Vina, VinaXB, and PLANTS were evaluated in the benchmarking study. All tools indicated better-than-random performance reflected by their pROC-AUC values against the Fascin crystal structure (PDB: ID 6I18). Interestingly, PLANTS exhibited the best screening performance and recognized potent actives at early enrichment. Accordingly, PLANTS was utilized in the prospective virtual screening effort for repurposing FDA-approved drugs (DrugBank database) and natural products (NANPDB). Further assessment via molecular dynamics simulations for 100 ns endorsed Remdesivir (DrugBank) and NANPDB3 (NANPDB) as potential binders to Fascin binding site 2. In conclusion, this study delivers a model for implementing a customized DEKOIS 2.0 benchmark set to enhance the VS success rate against new potential targets for cancer therapies.
Keywords: DEKOIS 2.0; Fascin; benchmarking; cancer; docking; virtual screening (VS).
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

