MtCLE08, MtCLE16, and MtCLE18 Transcription Patterns and Their Possible Functions in the Embryogenic Calli of Medicago truncatula
- PMID: 36771520
- PMCID: PMC9921462
- DOI: 10.3390/plants12030435
MtCLE08, MtCLE16, and MtCLE18 Transcription Patterns and Their Possible Functions in the Embryogenic Calli of Medicago truncatula
Abstract
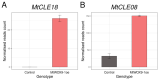
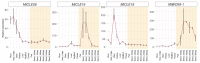
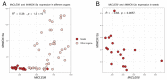
CLE peptides are well-known hormonal regulators of plant development, but their role in somatic embryogenesis remains undetermined. CLE genes are often regulated by WOX transcription factors and, in their turn, regulate the expression level of WOX genes. In this study, we used in vitro cultivation, as well as qPCR and transcriptomic analysis, to find CLE peptides which could regulate the MtWOX9-1 gene, stimulating somatic embryogenesis in Medicago truncatula. Three CLE peptides were found which could probably be such regulators, but none of them was found to influence MtWOX9-1 expression in the embryogenic calli. Nevertheless, overexpression of one of CLE genes under study, MtCLE16, decreased somatic embryogenesis intensity. Additionally, overexpression of MtCLE08 was found to suppress expression of MtWOX13a, a supposed antagonist of somatic embryo development. Our findings can be helpful for the search for new regeneration regulators which could be used for plant transformation.
Keywords: CLE peptides; Medicago truncatula; plant regeneration; somatic embryogenesis.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





Similar articles
-
MtWOX2 and MtWOX9-1 Effects on the Embryogenic Callus Transcriptome in Medicago truncatula.Plants (Basel). 2023 Dec 28;13(1):102. doi: 10.3390/plants13010102. Plants (Basel). 2023. PMID: 38202410 Free PMC article.
-
The WOX Genes from the Intermediate Clade: Influence on the Somatic Embryogenesis in Medicago truncatula.Plants (Basel). 2024 Jan 13;13(2):223. doi: 10.3390/plants13020223. Plants (Basel). 2024. PMID: 38256776 Free PMC article.
-
WUSCHEL Overexpression Promotes Callogenesis and Somatic Embryogenesis in Medicago truncatula Gaertn.Plants (Basel). 2021 Apr 7;10(4):715. doi: 10.3390/plants10040715. Plants (Basel). 2021. PMID: 33917135 Free PMC article.
-
WOX going on: CLE peptides in plant development.Curr Opin Plant Biol. 2021 Oct;63:102056. doi: 10.1016/j.pbi.2021.102056. Epub 2021 May 30. Curr Opin Plant Biol. 2021. PMID: 34077886 Free PMC article. Review.
-
Medicago truncatula transformation using leaf explants.Methods Mol Biol. 2006;343:115-27. doi: 10.1385/1-59745-130-4:115. Methods Mol Biol. 2006. PMID: 16988338 Review.
Cited by
-
MtWOX2 and MtWOX9-1 Effects on the Embryogenic Callus Transcriptome in Medicago truncatula.Plants (Basel). 2023 Dec 28;13(1):102. doi: 10.3390/plants13010102. Plants (Basel). 2023. PMID: 38202410 Free PMC article.
-
Functional Modules in the Meristems: "Tinkering" in Action.Plants (Basel). 2023 Oct 23;12(20):3661. doi: 10.3390/plants12203661. Plants (Basel). 2023. PMID: 37896124 Free PMC article. Review.
-
The WOX Genes from the Intermediate Clade: Influence on the Somatic Embryogenesis in Medicago truncatula.Plants (Basel). 2024 Jan 13;13(2):223. doi: 10.3390/plants13020223. Plants (Basel). 2024. PMID: 38256776 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

