An Interplay between Mitochondrial and ER Targeting of a Bacterial Signal Peptide in Plants
- PMID: 36771701
- PMCID: PMC9920398
- DOI: 10.3390/plants12030617
An Interplay between Mitochondrial and ER Targeting of a Bacterial Signal Peptide in Plants
Abstract
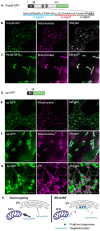
Protein targeting is essential in eukaryotic cells to maintain cell function and organelle identity. Signal peptides are a major type of targeting sequences containing a tripartite structure, which is conserved across all domains in life. They are frequently included in recombinant protein design in plants to increase yields by directing them to the endoplasmic reticulum (ER) or apoplast. The processing of bacterial signal peptides by plant cells is not well understood but could aid in the design of efficient heterologous expression systems. Here we analysed the signal peptide of the enzyme PmoB from methanotrophic bacteria. In plant cells, the PmoB signal peptide targeted proteins to both mitochondria and the ER. This dual localisation was still observed in a mutated version of the signal peptide sequence with enhanced mitochondrial targeting efficiency. Mitochondrial targeting was shown to be dependent on a hydrophobic region involved in transport to the ER. We, therefore, suggest that the dual localisation could be due to an ER-SURF pathway recently characterised in yeast. This work thus sheds light on the processing of bacterial signal peptides by plant cells and proposes a novel pathway for mitochondrial targeting in plants.
Keywords: ER-SURF; endoplasmic reticulum; mitochondria; plant cells; protein targeting; signal peptide; tobacco.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Barbante A., Irons S., Hawes C., Frigerio L., Vitale A., Pedrazzini E. Anchorage to the Cytosolic Face of the Endoplasmic Reticulum Membrane: A New Strategy to Stabilize a Cytosolic Recombinant Antigen in Plants. Plant Biotechnol. J. 2008;6:560–575. doi: 10.1111/j.1467-7652.2008.00342.x. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

