Novel Signposts on the Road from Natural Sources to Pharmaceutical Applications: A Combinative Approach between LC-DAD-MS and Offline LC-NMR for the Biochemical Characterization of Two Hypericum Species (H. montbretii and H. origanifolium)
- PMID: 36771732
- PMCID: PMC9921756
- DOI: 10.3390/plants12030648
Novel Signposts on the Road from Natural Sources to Pharmaceutical Applications: A Combinative Approach between LC-DAD-MS and Offline LC-NMR for the Biochemical Characterization of Two Hypericum Species (H. montbretii and H. origanifolium)
Abstract
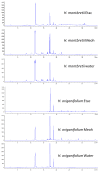
The members of the genus Hypericum have great potential to develop functional uses in nutraceutical and pharmaceutical applications. With this in mind, we aimed to determine the chemical profiling and biological properties of different extracts (ethyl acetate, methanol and water) from two Hypericum species (H. montbretii and H. origanifolium). We combined two approaches (LC-DAD-MS and LC-NMR) to identify and quantify chemical compounds of the extracts. Antioxidant properties (free radical quenching, reducing power and metal chelating) and enzyme inhibitory effects (cholinesterase, tyrosinase, amylase and glucosidase) were determined as biological properties. The tested extracts were rich in caffeic acid derivatives and flavonoids, and among them, 3-caffeoyl quinic acid and myricetin-3-O-rhamnoside were found to be the main compounds. The total phenolic and flavonoid levels were determined to be 50.97-134.99 mg GAE/g and 9.87-82.63 mg RE/g, respectively. With the exception of metal chelating, the methanol and water extracts showed stronger antioxidant properties than the ethyl acetate extracts. However, different results were obtained for each enzyme inhibition assay, and in general, the ethyl acetate extracts present more enzyme-inhibiting properties than the water or methanol extracts. Results from chemical and biological analyses were combined using multivariate analysis, which allowed establishing relationships between composition and observed effects of the Hypericum extracts based on the extraction solvents. To gain more insights between chemical compounds and enzyme-inhibiting effects, we performed molecular docking analysis. We observed favorable interactions between certain compounds and the tested enzymes during our analysis, confirming the data obtained from the multivariate approach. In conclusion, the obtained results may shed light on the road from natural sources to functional applications, and the tested Hypericum species may be considered potential raw materials, with promising chemical constituents and biological activities.
Keywords: Hypericum; bioactive agents; flavonoids; hydroxycinnamic acids; natural enzyme inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures











References
-
- Aware C.B., Patil D.N., Suryawanshi S.S., Mali P.R., Rane M.R., Gurav R.G., Jadhav J.P. Natural bioactive products as promising therapeutics: A review of natural product-based drug development. S. Afr. J. Bot. 2022;151:512–528. doi: 10.1016/j.sajb.2022.05.028. - DOI
-
- Li G., Lou M., Qi X. A brief overview of classical natural product drug synthesis and bioactivity. Org. Chem. Front. 2022;9:517–571. doi: 10.1039/D1QO01341F. - DOI
LinkOut - more resources
Full Text Sources

