Role of the histone tails in histone octamer transfer
- PMID: 36772826
- PMCID: PMC10164550
- DOI: 10.1093/nar/gkad079
Role of the histone tails in histone octamer transfer
Abstract
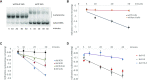
The exceptionally high positive charge of the histones, concentrated in the N- and C-terminal tails, is believed to contribute to the stability of the nucleosome by neutralizing the negative charge of the nucleosomal DNA. We find, on the contrary, that the high positive charge contributes to instability, performing an essential function in chromatin remodeling. We show that the tails are required for removal of the histone octamer by the RSC chromatin remodeling complex, and this function is not due to direct RSC-tail interaction. We also show that the tails are required for histone octamer transfer from nucleosomes to DNA, and this activity of the tails is a consequence of their positive charge. Thus, the histone tails, intrinsically disordered protein regions, perform a critical role in chromatin structure and transcription, unrelated to their well-known role in regulation through posttranscriptional modification.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Kornberg R.D., Lorch Y.. Primary role of the nucleosome. Mol. Cell. 2020; 79:371–375. - PubMed
-
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997; 389:251–260. - PubMed
-
- Lorch Y., Kornberg R.D.. Isolation and assay of the RSC chromatin-remodeling complex from Saccharomyces cerevisiae. Methods Enzymol. 2003; 377:316–322. - PubMed
-
- Wittmeyer J., Saha A., Cairns B.. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 2004; 377:322–343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

