Characterisation of SARS-CoV-2 variants in Beijing during 2022: an epidemiological and phylogenetic analysis
- PMID: 36773619
- PMCID: PMC9949854
- DOI: 10.1016/S0140-6736(23)00129-0
Characterisation of SARS-CoV-2 variants in Beijing during 2022: an epidemiological and phylogenetic analysis
Abstract
Background: Due to the national dynamic zero-COVID strategy in China, there were no persistent local transmissions of SARS-CoV-2 in Beijing before December, 2022. However, imported cases have been frequently detected over the past 3 years. With soaring growth in the number of COVID-19 cases in China recently, there are concerns that there might be an emergence of novel SARS-CoV-2 variants. Routine surveillance of viral genomes has been carried out in Beijing over the last 3 years. Spatiotemporal analyses of recent viral genome sequences compared with that of global pooled and local data are crucial for the global response to the ongoing COVID-19 pandemic.
Methods: We routinely collected respiratory samples covering both imported and local cases in Beijing for the last 3 years (of which the present study pertains to samples collected between January and December, 2022), and then randomly selected samples for analysis. Next-generation sequencing was used to generate the SARS-CoV-2 genomes. Phylogenetic and population dynamic analyses were performed using high-quality complete sequences in this study.
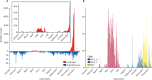
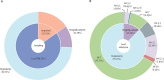
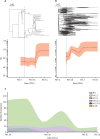
Findings: We obtained a total of 2994 complete SARS-CoV-2 genome sequences in this study, among which 2881 were high quality and were used for further analysis. From Nov 14 to Dec 20, we sequenced 413 new samples, including 350 local cases and 63 imported cases. All of these genomes belong to the existing 123 Pango lineages, showing there are no persistently dominant variants or novel lineages. Nevertheless, BA.5.2 and BF.7 are currently dominant in Beijing, accounting for 90% of local cases since Nov 14 (315 of 350 local cases sequenced in this study). The effective population size for both BA.5.2 and BF.7 in Beijing increased after Nov 14, 2022.
Interpretation: The co-circulation of BF.7 and BA.5.2 has been present in the current outbreak since Nov 14, 2022 in Beijing, and there is no evidence that novel variants emerged. Although our data were only from Beijing, the results could be considered a snapshot of China, due to the frequent population exchange and the presence of circulating strains with high transmissibility.
Funding: National Key Research and Development Program of China and Strategic Priority Research Program of the Chinese Academy of Sciences.
Translation: For the Chinese translation of the abstract see Supplementary Materials section.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests We declare no competing interests.
Figures

Comment in
-
So far, no novel SARS-CoV-2 variants from Beijing-and hopefully better scientific cooperation going forward.Lancet. 2023 Feb 25;401(10377):621-622. doi: 10.1016/S0140-6736(23)00268-4. Epub 2023 Feb 8. Lancet. 2023. PMID: 36773617 Free PMC article. No abstract available.
-
Interpreting COVID-19 data from China: a call for caution.Lancet. 2024 Jan 13;403(10422):144. doi: 10.1016/S0140-6736(23)01745-2. Lancet. 2024. PMID: 38218609 No abstract available.
References
-
- WHO Tracking SARS-CoV-2 variants. https://www.who.int/activities/tracking-SARS-CoV-2-variants
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

