Genomic and microenvironmental heterogeneity shaping epithelial-to-mesenchymal trajectories in cancer
- PMID: 36774358
- PMCID: PMC9922305
- DOI: 10.1038/s41467-023-36439-7
Genomic and microenvironmental heterogeneity shaping epithelial-to-mesenchymal trajectories in cancer
Abstract
The epithelial to mesenchymal transition (EMT) is a key cellular process underlying cancer progression, with multiple intermediate states whose molecular hallmarks remain poorly characterised. To fill this gap, we present a method to robustly evaluate EMT transformation in individual tumours based on transcriptomic signals. We apply this approach to explore EMT trajectories in 7180 tumours of epithelial origin and identify three macro-states with prognostic and therapeutic value, attributable to epithelial, hybrid E/M and mesenchymal phenotypes. We show that the hybrid state is relatively stable and linked with increased aneuploidy. We further employ spatial transcriptomics and single cell datasets to explore the spatial heterogeneity of EMT transformation and distinct interaction patterns with cytotoxic, NK cells and fibroblasts in the tumour microenvironment. Additionally, we provide a catalogue of genomic events underlying distinct evolutionary constraints on EMT transformation. This study sheds light on the aetiology of distinct stages along the EMT trajectory, and highlights broader genomic and environmental hallmarks shaping the mesenchymal transformation of primary tumours.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

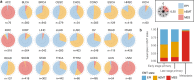



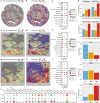


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

