Low muscle mass in lung cancer is associated with an inflammatory and immunosuppressive tumor microenvironment
- PMID: 36774484
- PMCID: PMC9921698
- DOI: 10.1186/s12967-023-03901-5
Low muscle mass in lung cancer is associated with an inflammatory and immunosuppressive tumor microenvironment
Abstract
Background: Computed tomographies (CT) are useful for identifying muscle loss in non-small lung cancer (NSCLC) cachectic patients. However, we lack consensus on the best cutoff point for pectoralis muscle loss. We aimed to characterize NSCLC patients based on muscularity, clinical data, and the transcriptional profile from the tumor microenvironment to build a cachexia classification model.
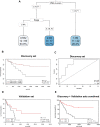
Methods: We used machine learning to generate a muscle loss prediction model, and the tumor's cellular and transcriptional profile was characterized in patients with low muscularity. First, we measured the pectoralis muscle area (PMA) of 211 treatment-naive NSCLC patients using CT available in The Cancer Imaging Archive. The cutoffs were established using machine learning algorithms (CART and Cutoff Finder) on PMA, clinical, and survival data. We evaluated the prediction model in a validation set (36 NSCLC). Tumor RNA-Seq (GSE103584) was used to profile the transcriptome and cellular composition based on digital cytometry.
Results: CART demonstrated that a lower PMA was associated with a high risk of death (HR = 1.99). Cutoff Finder selected PMA cutoffs separating low-muscularity (LM) patients based on the risk of death (P-value = 0.003; discovery set). The cutoff presented 84% of success in classifying low muscle mass. The high risk of LM patients was also found in the validation set. Tumor RNA-Seq revealed 90 upregulated secretory genes in LM that potentially interact with muscle cell receptors. The LM upregulated genes enriched inflammatory biological processes. Digital cytometry revealed that LM patients presented high proportions of cytotoxic and exhausted CD8+ T cells.
Conclusions: Our prediction model identified cutoffs that distinguished patients with lower PMA and survival with an inflammatory and immunosuppressive TME enriched with inflammatory factors and CD8+ T cells.
Keywords: CD8+ T cells; Computed tomography; Machine learning; Non-small cell lung cancer; Transcriptomics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

