Regional and cell-type-specific afferent and efferent projections of the mouse claustrum
- PMID: 36774552
- PMCID: PMC10415534
- DOI: 10.1016/j.celrep.2023.112118
Regional and cell-type-specific afferent and efferent projections of the mouse claustrum
Abstract
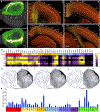
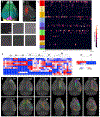
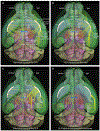
The claustrum (CLA) is a conspicuous subcortical structure interconnected with cortical and subcortical regions. Its regional anatomy and cell-type-specific connections in the mouse remain not fully determined. Using multimodal reference datasets, we confirmed the delineation of the mouse CLA as a single group of neurons embedded in the agranular insular cortex. We quantitatively investigated brain-wide inputs and outputs of CLA using bulk anterograde and retrograde viral tracing data and single neuron tracing data. We found that the prefrontal module has more cell types projecting to the CLA than other cortical modules, with layer 5 IT neurons predominating. We found nine morphological types of CLA principal neurons that topographically innervate functionally linked cortical targets, preferentially the midline cortical areas, secondary motor area, and entorhinal area. Together, this study provides a detailed wiring diagram of the cell-type-specific connections of the mouse CLA, laying a foundation for studying its functions at the cellular level.
Keywords: AAV; CP: Neuroscience; anterograde tracing; cell type; claustrum; isocortex; mouse; rabies virus; retrograde tracing; single neuron reconstruction.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Bruguier H, Suarez R, Manger P, Hoerder-Suabedissen A, Shelton AM, Oliver DK, Packer AM, Ferran JL, García-Moreno F, Puelles L, and Molnár Z (2020). In search of common developmental and evolutionary origin of the claustrum and subplate. J. Comp. Neurol. 528, 2956–2977. 10.1002/cne.24922. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

