MARS2 drives metabolic switch of non-small-cell lung cancer cells via interaction with MCU
- PMID: 36774778
- PMCID: PMC9947422
- DOI: 10.1016/j.redox.2023.102628
MARS2 drives metabolic switch of non-small-cell lung cancer cells via interaction with MCU
Abstract
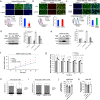
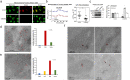
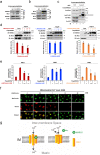
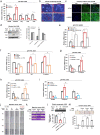
Mitochondrial methionyl-tRNA synthetase (MARS2) canonically mediates the formation of fMet-tRNAifMet for mitochondrial translation initiation. Mitochondrial calcium uniporter (MCU) is a major gate of Ca2+ flux from cytosol into the mitochondrial matrix. We found that MARS2 interacts with MCU and stimulates mitochondrial Ca2+ influx. Methionine binding to MARS2 would act as a molecular switch that regulates MARS2-MCU interaction. Endogenous knockdown of MARS2 attenuates mitochondrial Ca2+ influx and induces p53 upregulation through the Ca2+-dependent CaMKII/CREB signaling. Subsequently, metabolic rewiring from glycolysis into pentose phosphate pathway is triggered and cellular reactive oxygen species level decreases. This metabolic switch induces inhibition of epithelial-mesenchymal transition (EMT) via cellular redox regulation. Expression of MARS2 is regulated by ZEB1 transcription factor in response to Wnt signaling. Our results suggest the mechanisms of mitochondrial Ca2+ uptake and metabolic control of cancer that are exerted by the key factors of the mitochondrial translational machinery and Ca2+ homeostasis.
Keywords: Cancer metabolism; Epithelial-mesenchymal transition; Mitochondrial calcium uniporter; Mitochondrial methionyl-tRNA synthetase; Reactive oxygen species; p53.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

