Single-molecule dynamics of DNA gyrase in evolutionarily distant bacteria Mycobacterium tuberculosis and Escherichia coli
- PMID: 36775125
- PMCID: PMC10130225
- DOI: 10.1016/j.jbc.2023.103003
Single-molecule dynamics of DNA gyrase in evolutionarily distant bacteria Mycobacterium tuberculosis and Escherichia coli
Abstract
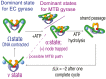
DNA gyrase is an essential nucleoprotein motor present in all bacteria and is a major target for antibiotic treatment of Mycobacterium tuberculosis (MTB) infection. Gyrase hydrolyzes ATP to add negative supercoils to DNA using a strand passage mechanism that has been investigated using biophysical and biochemical approaches. To analyze the dynamics of substeps leading to strand passage, single-molecule rotor bead tracking (RBT) has been used previously to follow real-time supercoiling and conformational transitions in Escherichia coli (EC) gyrase. However, RBT has not yet been applied to gyrase from other pathogenically relevant bacteria, and it is not known whether substeps are conserved across evolutionarily distant species. Here, we compare gyrase supercoiling dynamics between two evolutionarily distant bacterial species, MTB and EC. We used RBT to measure supercoiling rates, processivities, and the geometries and transition kinetics of conformational states of purified gyrase proteins in complex with DNA. Our results show that E. coli and MTB gyrases are both processive, with the MTB enzyme displaying velocities ∼5.5× slower than the EC enzyme. Compared with EC gyrase, MTB gyrase also more readily populates an intermediate state with DNA chirally wrapped around the enzyme, in both the presence and absence of ATP. Our substep measurements reveal common features in conformational states of EC and MTB gyrases interacting with DNA but also suggest differences in populations and transition rates that may reflect distinct cellular needs between these two species.
Keywords: DNA-protein interaction; bacterial genetics; bacterial pathogenesis; bacterial transcription; molecular motor; single-molecule biophysics; topoisomerase.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Forterre P., Gribaldo S., Gadelle D., Serre M.-C. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. - PubMed
-
- Van Boeckel T.P., Gandra S., Ashok A., Caudron Q., Grenfell B.T., Levin S.A., et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014;14:742–750. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

