Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease
- PMID: 36775130
- PMCID: PMC9916189
- DOI: 10.1016/j.jbc.2023.103004
Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease
Abstract
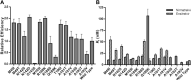
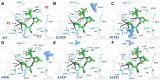
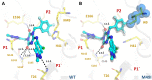
SARS-CoV-2 is the causative agent of COVID-19. The main viral protease (Mpro) is an attractive target for antivirals. The clinically approved drug nirmatrelvir and the clinical candidate ensitrelvir have so far showed great potential for treatment of viral infection. However, the broad use of antivirals is often associated with resistance generation. Herein, we enzymatically characterized 14 naturally occurring Mpro polymorphisms that are close to the binding site of these antivirals. Nirmatrelvir retained its potency against most polymorphisms tested, while mutants G143S and Q189K were associated with diminished inhibition constants. For ensitrelvir, diminished inhibition constants were observed for polymorphisms M49I, G143S, and R188S, but not for Q189K, suggesting a distinct resistance profile between inhibitors. In addition, the crystal structures of selected polymorphisms revealed interactions that were critical for loss of potency. In conclusion, our data will assist the monitoring of potential resistant strains, support the design of combined therapy, as well as assist the development of the next generation of Mpro inhibitors.
Keywords: COVID; SARS-CoV-2; drug; ensitrelvir; main protease; mpro; mutation; nirmatrelvir; paxlovid; resistance; structure; x-ray; xocova.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. - PubMed
-
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., et al. The coding capacity of SARS-CoV-2. Nature. 2020;589:125–130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

