Allelic variations of Vrn-1 and Ppd-1 genes in Japanese wheat varieties reveal the genotype-environment interaction for heading time
- PMID: 36776445
- PMCID: PMC9895800
- DOI: 10.1270/jsbbs.22017
Allelic variations of Vrn-1 and Ppd-1 genes in Japanese wheat varieties reveal the genotype-environment interaction for heading time
Abstract
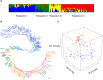
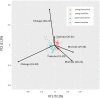
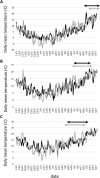
The timing of heading is largely affected by environmental conditions. In wheat, Vrn-1 and Ppd-1 have been identified as the major genes involved in vernalization requirement and photoperiod sensitivity, respectively. To compare the effects of Vrn-1 and Ppd-1 alleles on heading time under different environments, we genotyped Vrn-1 and Ppd-1 homoeologues and measured the heading time at Morioka, Tsukuba and Chikugo in Japan for two growing seasons. A total of 128 Japanese and six foreign varieties, classified into four populations based on the 519 genome-wide SNPs, were used for analysis. Varieties with the spring alleles (Vrn-D1a or Vrn-D1b) at the Vrn-D1 locus and insensitive allele (Hapl-I) at the Ppd-D1 locus were found in earlier heading varieties. The effects of Vrn-D1 and Ppd-D1 on heading time were stronger than those of the other Vrn-1 and Ppd-1 homoeologues. Analysis of variance revealed that heading time was significantly affected by the genotype-environment interactions. Some Vrn-1 and Ppd-1 alleles conferred earlier or later heading in specific environments, indicating that the effect of both alleles on the timing of heading depends on the environment. Information on Vrn-1 and Ppd-1 alleles, together with heading time in various environments, provide useful information for wheat breeding.
Keywords: Ppd-1; Vrn-1; genotype-environment interaction; heading time; wheat.
Copyright © 2022 by JAPANESE SOCIETY OF BREEDING.
Figures


Similar articles
-
Genetics of flowering time in bread wheat Triticum aestivum: complementary interaction between vernalization-insensitive and photoperiod-insensitive mutations imparts very early flowering habit to spring wheat.J Genet. 2012;91(1):33-47. doi: 10.1007/s12041-012-0149-3. J Genet. 2012. PMID: 22546824
-
Allelic variation at the vernalization and photoperiod sensitivity loci in Chinese winter wheat cultivars (Triticum aestivum L.).Front Plant Sci. 2015 Jul 1;6:470. doi: 10.3389/fpls.2015.00470. eCollection 2015. Front Plant Sci. 2015. PMID: 26191066 Free PMC article.
-
Interactive effects of multiple vernalization (Vrn-1)- and photoperiod (Ppd-1)-related genes on the growth habit of bread wheat and their association with heading and flowering time.BMC Plant Biol. 2018 Dec 27;18(1):374. doi: 10.1186/s12870-018-1587-8. BMC Plant Biol. 2018. PMID: 30587132 Free PMC article.
-
Genome-wide association study of heading and flowering dates and construction of its prediction equation in Chinese common wheat.Theor Appl Genet. 2018 Nov;131(11):2271-2285. doi: 10.1007/s00122-018-3181-8. Epub 2018 Sep 14. Theor Appl Genet. 2018. PMID: 30218294 Review.
-
Allelic Variations in Vernalization (Vrn) Genes in Triticum spp.Genes (Basel). 2024 Feb 17;15(2):251. doi: 10.3390/genes15020251. Genes (Basel). 2024. PMID: 38397240 Free PMC article. Review.
Cited by
-
Identification and validation of a major quantitative trait locus for precise control of heading date in wheat (Triticum aestivum L.).BMC Plant Biol. 2025 May 10;25(1):616. doi: 10.1186/s12870-025-06646-z. BMC Plant Biol. 2025. PMID: 40348961 Free PMC article.
-
Identification of the causal mutation in early heading mutant of bread wheat (Triticum aestivum L.) using MutMap approach.Mol Breed. 2024 May 21;44(6):41. doi: 10.1007/s11032-024-01478-5. eCollection 2024 Jun. Mol Breed. 2024. PMID: 38779634 Free PMC article.
-
Natural variations of wheat EARLY FLOWERING 3 highlight their contributions to local adaptation through fine-tuning of heading time.Theor Appl Genet. 2023 May 26;136(6):139. doi: 10.1007/s00122-023-04386-y. Theor Appl Genet. 2023. PMID: 37233781
References
-
- Beales, J., Turner A., Griffiths S., Snape J.W. and Laurie D.A. (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115: 721–733. - PubMed
-
- Bennett, D., Izanloo A., Edwards J., Kuchel H., Chalmers K., Tester M., Reynolds M., Schnurbusch T. and Langridge P. (2012) Identification of novel quantitative trait loci for days to ear emergence and flag leaf glaucousness in a bread wheat (Triticum aestivum L.) population adapted to southern Australian conditions. Theor Appl Genet 124: 697–711. - PubMed

