A targeted multi-proteomics approach generates a blueprint of the ciliary ubiquitinome
- PMID: 36776558
- PMCID: PMC9908615
- DOI: 10.3389/fcell.2023.1113656
A targeted multi-proteomics approach generates a blueprint of the ciliary ubiquitinome
Abstract
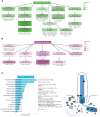
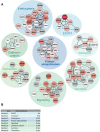
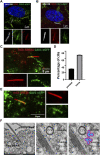
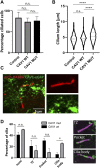
Establishment and maintenance of the primary cilium as a signaling-competent organelle requires a high degree of fine tuning, which is at least in part achieved by a variety of post-translational modifications. One such modification is ubiquitination. The small and highly conserved ubiquitin protein possesses a unique versatility in regulating protein function via its ability to build mono and polyubiquitin chains onto target proteins. We aimed to take an unbiased approach to generate a comprehensive blueprint of the ciliary ubiquitinome by deploying a multi-proteomics approach using both ciliary-targeted ubiquitin affinity proteomics, as well as ubiquitin-binding domain-based proximity labelling in two different mammalian cell lines. This resulted in the identification of several key proteins involved in signaling, cytoskeletal remodeling and membrane and protein trafficking. Interestingly, using two different approaches in IMCD3 and RPE1 cells, respectively, we uncovered several novel mechanisms that regulate cilia function. In our IMCD3 proximity labeling cell line model, we found a highly enriched group of ESCRT-dependent clathrin-mediated endocytosis-related proteins, suggesting an important and novel role for this pathway in the regulation of ciliary homeostasis and function. In contrast, in RPE1 cells we found that several structural components of caveolae (CAV1, CAVIN1, and EHD2) were highly enriched in our cilia affinity proteomics screen. Consistently, the presence of caveolae at the ciliary pocket and ubiquitination of CAV1 specifically, were found likely to play a role in the regulation of ciliary length in these cells. Cilia length measurements demonstrated increased ciliary length in RPE1 cells stably expressing a ubiquitination impaired CAV1 mutant protein. Furthermore, live cell imaging in the same cells revealed decreased CAV1 protein turnover at the cilium as the possible cause for this phenotype. In conclusion, we have generated a comprehensive list of cilia-specific proteins that are subject to regulation via ubiquitination which can serve to further our understanding of cilia biology in health and disease.
Keywords: ESCRT; caveolae; cilia; cilia ubiquitination; ciliary proteostasis; ciliopathies; clathrin-mediated endocytosis.
Copyright © 2023 Aslanyan, Doornbos, Diwan, Anvarian, Beyer, Junger, van Beersum, Russell, Ueffing, Ludwig, Boldt, Pedersen and Roepman.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Alexa A., Rahnenfuhrer J. (2020). topGO: Enrichment analysis for gene ontology.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

