Integrated landscape of cardiac metabolism in end-stage human nonischemic dilated cardiomyopathy
- PMID: 36776621
- PMCID: PMC9910091
- DOI: 10.1038/s44161-022-00117-6
Integrated landscape of cardiac metabolism in end-stage human nonischemic dilated cardiomyopathy
Abstract
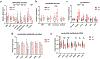
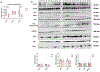
Heart failure (HF) is a leading cause of mortality. Failing hearts undergo profound metabolic changes, but a comprehensive evaluation in humans is lacking. We integrate plasma and cardiac tissue metabolomics of 678 metabolites, genome-wide RNA-sequencing, and proteomic studies to examine metabolic status in 87 explanted human hearts from 39 patients with end-stage HF compared with 48 nonfailing donors. We confirm bioenergetic defects in human HF and reveal selective depletion of adenylate purines required for maintaining ATP levels. We observe substantial reductions in fatty acids and acylcarnitines in failing tissue, despite plasma elevations, suggesting defective import of fatty acids into cardiomyocytes. Glucose levels, in contrast, are elevated. Pyruvate dehydrogenase, which gates carbohydrate oxidation, is de-repressed, allowing increased lactate and pyruvate burning. Tricarboxylic acid cycle intermediates are significantly reduced. Finally, bioactive lipids are profoundly reprogrammed, with marked reductions in ceramides and elevations in lysoglycerophospholipids. These data unveil profound metabolic abnormalities in human failing hearts.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures








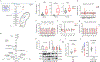

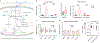

References
-
- Shah KS et al. Heart failure with preserved, borderline, and reduced ejection fraction: 5-year outcomes. J. Am. Coll. Cardiol 70, 2476–2486 (2017). - PubMed
-
- Ingwall JS & Weiss RG Is the failing heart energy starved? On using chemical energy to support cardiac function. Circ. Res 95, 135–145 (2004). - PubMed
-
- Neubauer S The failing heart—an engine out of fuel. N. Engl. J. Med 356, 1140–1151 (2007). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
