Network analyses of upper and lower airway transcriptomes identify shared mechanisms among children with recurrent wheezing and school-age asthma
- PMID: 36776870
- PMCID: PMC9911682
- DOI: 10.3389/fimmu.2023.1087551
Network analyses of upper and lower airway transcriptomes identify shared mechanisms among children with recurrent wheezing and school-age asthma
Abstract
Background: Predicting which preschool children with recurrent wheezing (RW) will develop school-age asthma (SA) is difficult, highlighting the critical need to clarify the pathogenesis of RW and the mechanistic relationship between RW and SA. Despite shared environmental exposures and genetic determinants, RW and SA are usually studied in isolation. Based on network analysis of nasal and tracheal transcriptomes, we aimed to identify convergent transcriptomic mechanisms in RW and SA.
Methods: RNA-sequencing data from nasal and tracheal brushing samples were acquired from the Gene Expression Omnibus. Combined with single-cell transcriptome data, cell deconvolution was used to infer the composition of 18 cellular components within the airway. Consensus weighted gene co-expression network analysis was performed to identify consensus modules closely related to both RW and SA. Shared pathways underlying consensus modules between RW and SA were explored by enrichment analysis. Hub genes between RW and SA were identified using machine learning strategies and validated using external datasets and quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Finally, the potential value of hub genes in defining RW subsets was determined using nasal and tracheal transcriptome data.
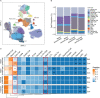
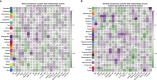
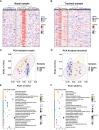
Results: Co-expression network analysis revealed similarities in the transcriptional networks of RW and SA in the upper and lower airways. Cell deconvolution analysis revealed an increase in mast cell fraction but decrease in club cell fraction in both RW and SA airways compared to controls. Consensus network analysis identified two consensus modules highly associated with both RW and SA. Enrichment analysis of the two consensus modules indicated that fatty acid metabolism-related pathways were shared key signals between RW and SA. Furthermore, machine learning strategies identified five hub genes, i.e., CST1, CST2, CST4, POSTN, and NRTK2, with the up-regulated hub genes in RW and SA validated using three independent external datasets and qRT-PCR. The gene signatures of the five hub genes could potentially be used to determine type 2 (T2)-high and T2-low subsets in preschoolers with RW.
Conclusions: These findings improve our understanding of the molecular pathogenesis of RW and provide a rationale for future exploration of the mechanistic relationship between RW and SA.
Keywords: RNA-seq; cell deconvolution; gene co-expression network; machine learning; recurrent wheezing; scRNA-seq; school-age asthma.
Copyright © 2023 Wang, He, Li, Zhao, Zhang and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Identification of surrogate prognostic biomarkers for allergic asthma in nasal epithelial brushing samples by WGCNA.J Cell Biochem. 2019 Apr;120(4):5137-5150. doi: 10.1002/jcb.27790. Epub 2018 Oct 10. J Cell Biochem. 2019. PMID: 30304558
-
Identification of Molecular Markers Related to Immune Infiltration in Patients with Severe Asthma: A Comprehensive Bioinformatics Analysis Based on the Human Bronchial Epithelial Transcriptome.Dis Markers. 2022 Nov 3;2022:8906064. doi: 10.1155/2022/8906064. eCollection 2022. Dis Markers. 2022. PMID: 36393974 Free PMC article.
-
Transcriptomic analysis identified SLC40A1 as a key iron metabolism-related gene in airway macrophages in childhood allergic asthma.Front Cell Dev Biol. 2023 Apr 13;11:1164544. doi: 10.3389/fcell.2023.1164544. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37123407 Free PMC article.
-
Oropharyngeal Microbiota Clusters in Children with Asthma or Wheeze Associate with Allergy, Blood Transcriptomic Immune Pathways, and Exacerbation Risk.Am J Respir Crit Care Med. 2023 Jul 15;208(2):142-154. doi: 10.1164/rccm.202211-2107OC. Am J Respir Crit Care Med. 2023. PMID: 37163754
-
Low-grade disease activity in early life precedes childhood asthma and allergy.Dan Med J. 2016 Aug;63(8):B5272. Dan Med J. 2016. PMID: 27477800 Review.
Cited by
-
Extracellular vesicles from bronchoalveolar lavage fluid provide insights into the inhaled corticosteroids treatment response in COPD.Respir Res. 2025 Jul 30;26(1):254. doi: 10.1186/s12931-025-03330-6. Respir Res. 2025. PMID: 40739568 Free PMC article. Clinical Trial.
-
Salidroside sensitizes Triple-negative breast cancer to ferroptosis by SCD1-mediated lipogenesis and NCOA4-mediated ferritinophagy.J Adv Res. 2025 Aug;74:589-607. doi: 10.1016/j.jare.2024.09.027. Epub 2024 Sep 29. J Adv Res. 2025. PMID: 39353532 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

