Stimulation of the farnesoid X receptor promotes M2 macrophage polarization
- PMID: 36776885
- PMCID: PMC9911659
- DOI: 10.3389/fimmu.2023.1065790
Stimulation of the farnesoid X receptor promotes M2 macrophage polarization
Abstract
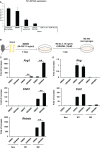
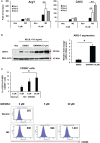
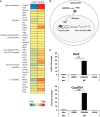
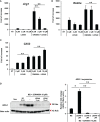
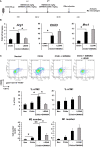
FXR is a key molecule that modulates anti-inflammatory activity in the intestinal-liver axis. Although FXR has pleiotropic functions including regulation of liver inflammation and activation of macrophages, it remains unclear whether it is involved in macrophage polarization. In this paper we demonstrated that stimulation of macrophages derived from the bone marrow using an FXR agonist activated polarization toward M2 but not M1 macrophages. The treatment of mice with chitin skewed macrophage polarization towards M2 macrophages, while co-treatment with an FXR agonist further promoted the polarization toward M2 macrophages in vivo. This skewed polarization towards M2 macrophages by an FXR agonist was accompanied by increased expression of signaling molecules related to the retinoic acid receptor. Inhibition of the retinoic acid receptor suppressed FXR agonist-mediated M2 macrophage polarization, indicating that this polarization was, at least, partly dependent on the retinoic acid receptor pathway. These data demonstrate that FXR has a role in polarization toward M2 macrophages and suggest a possible therapeutic potential of FXR agonists in M2 macrophage-related conditions.
Keywords: cell differentiation; farnesoid X receptor; inflammation; macrophage; tissue repair.
Copyright © 2023 Jaroonwitchawan, Arimochi, Sasaki, Ishifune, Kondo, Otsuka, Tsukumo and Yasutomo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Bile Acids Elevated in Chronic Periaortitis Could Activate Farnesoid-X-Receptor to Suppress IL-6 Production by Macrophages.Front Immunol. 2021 Apr 22;12:632864. doi: 10.3389/fimmu.2021.632864. eCollection 2021. Front Immunol. 2021. PMID: 33968024 Free PMC article.
-
FXR agonist GW4064 alleviates endotoxin-induced hepatic inflammation by repressing macrophage activation.World J Gastroenterol. 2014 Oct 21;20(39):14430-41. doi: 10.3748/wjg.v20.i39.14430. World J Gastroenterol. 2014. PMID: 25339829 Free PMC article.
-
LncGBP9/miR-34a axis drives macrophages toward a phenotype conducive for spinal cord injury repair via STAT1/STAT6 and SOCS3.J Neuroinflammation. 2020 Apr 28;17(1):134. doi: 10.1186/s12974-020-01805-5. J Neuroinflammation. 2020. PMID: 32345320 Free PMC article.
-
Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways.Cell Signal. 2014 Feb;26(2):192-7. doi: 10.1016/j.cellsig.2013.11.004. Epub 2013 Nov 9. Cell Signal. 2014. PMID: 24219909 Review.
-
Forkhead Box O1 Regulates Macrophage Polarization Following Staphylococcus aureus Infection: Experimental Murine Data and Review of the Literature.Clin Rev Allergy Immunol. 2016 Dec;51(3):353-369. doi: 10.1007/s12016-016-8531-1. Clin Rev Allergy Immunol. 2016. PMID: 26924010 Review.
Cited by
-
Yinchenhao Decoction Protects Against Acute Liver Injury in Mice With Biliary Acute Pancreatitis by Regulating the Gut Microflora-Bile Acids-Liver Axis.Gastroenterol Res Pract. 2024 Jun 27;2024:8882667. doi: 10.1155/2024/8882667. eCollection 2024. Gastroenterol Res Pract. 2024. PMID: 38966598 Free PMC article.
-
Spatial and phenotypic heterogeneity of resident and monocyte-derived macrophages during inflammatory exacerbations leading to pulmonary fibrosis.Front Immunol. 2024 Jul 19;15:1425466. doi: 10.3389/fimmu.2024.1425466. eCollection 2024. Front Immunol. 2024. PMID: 39100672 Free PMC article.
-
FXR activation reduces the formation of macrophage foam cells and atherosclerotic plaque, possibly by down regulating hepatic lipase in macrophages.FEBS Open Bio. 2025 Feb;15(2):311-323. doi: 10.1002/2211-5463.13925. Epub 2024 Nov 27. FEBS Open Bio. 2025. PMID: 39601316 Free PMC article.
-
Impact of intestinal microenvironments in obesity and bariatric surgery on shaping macrophages.Immunometabolism (Cobham). 2023 Nov 28;5(4):e00033. doi: 10.1097/IN9.0000000000000033. eCollection 2023 Oct. Immunometabolism (Cobham). 2023. PMID: 38037591 Free PMC article. Review.
-
Targeting Farnesoid X Receptor in Tumor and the Tumor Microenvironment: Implication for Therapy.Int J Mol Sci. 2023 Dec 19;25(1):6. doi: 10.3390/ijms25010006. Int J Mol Sci. 2023. PMID: 38203175 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

