Identification of potential biomarkers and immune infiltration characteristics in recurrent implantation failure using bioinformatics analysis
- PMID: 36776897
- PMCID: PMC9909740
- DOI: 10.3389/fimmu.2023.992765
Identification of potential biomarkers and immune infiltration characteristics in recurrent implantation failure using bioinformatics analysis
Abstract
Introduction: Recurrent implantation failure (RIF) is a frustrating challenge because the cause is unknown. The current study aims to identify differentially expressed genes (DEGs) in the endometrium on the basis of immune cell infiltration characteristics between RIF patients and healthy controls, as well as to investigate potential prognostic markers in RIF.
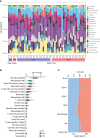
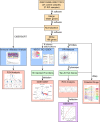
Methods: GSE103465, and GSE111974 datasets from the Gene Expression Omnibus database were obtained to screen DEGs between RIF and control groups. Gene Ontology analysis, Kyoto Encyclopedia of Genes and Genomes Pathway analysis, Gene Set Enrichment Analysis, and Protein-protein interactions analysis were performed to investigate potential biological functions and signaling pathways. CIBERSORT was used to describe the level of immune infiltration in RIF, and flow cytometry was used to confirm the top two most abundant immune cells detected.
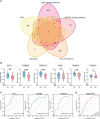
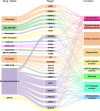
Results: 122 downregulated and 66 upregulated DEGs were obtained between RIF and control groups. Six immune-related hub genes were discovered, which were involved in Wnt/-catenin signaling and Notch signaling as a result of our research. The ROC curves revealed that three of the six identified genes (AKT1, PSMB8, and PSMD10) had potential diagnostic values for RIF. Finally, we used cMap analysis to identify potential therapeutic or induced compounds for RIF, among which fulvestrant (estrogen receptor antagonist), bisindolylmaleimide-ix (CDK and PKC inhibitor), and JNK-9L (JNK inhibitor) were thought to influence the pathogenic process of RIF. Furthermore, our findings revealed the level of immune infiltration in RIF by highlighting three signaling pathways (Wnt/-catenin signaling, Notch signaling, and immune response) and three potential diagnostic DEGs (AKT1, PSMB8, and PSMD10).
Conclusion: Importantly, our findings may contribute to the scientific basis for several potential therapeutic agents to improve endometrial receptivity.
Keywords: Wnt/β-catenin signaling; bioinformatics; biomarkers; immune infiltration; notch signaling; recurrent implantation failure.
Copyright © 2023 Lai, Zhang, Zhou, Shi, Yang, Yang, Wu, Xie, Zhang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest. The reviewer JZ declared a shared parent affiliation with the author WJZ to the handling editor at the time of the review.
Figures

Similar articles
-
Analysis of weighted gene co-expression networks and clinical validation identify hub genes and immune cell infiltration in the endometrial cells of patients with recurrent implantation failure.Front Genet. 2024 Apr 5;15:1292757. doi: 10.3389/fgene.2024.1292757. eCollection 2024. Front Genet. 2024. PMID: 38645487 Free PMC article.
-
Shared diagnostic biomarkers and underlying mechanisms between endometriosis and recurrent implantation failure.Front Endocrinol (Lausanne). 2025 Feb 19;16:1490746. doi: 10.3389/fendo.2025.1490746. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40046872 Free PMC article.
-
Gene profiling reveals the role of inflammation, abnormal uterine muscle contraction and vascularity in recurrent implantation failure.Front Genet. 2023 Feb 24;14:1108805. doi: 10.3389/fgene.2023.1108805. eCollection 2023. Front Genet. 2023. PMID: 36911409 Free PMC article.
-
Identification of key genes and immune cell infiltration in recurrent implantation failure: A study based on integrated analysis of multiple microarray studies.Am J Reprod Immunol. 2022 Oct;88(4):e13607. doi: 10.1111/aji.13607. Epub 2022 Aug 15. Am J Reprod Immunol. 2022. PMID: 35929523 Free PMC article.
-
Immunomodulation for unexplained recurrent implantation failure: where are we now?Reproduction. 2023 Jan 4;165(2):R39-R60. doi: 10.1530/REP-22-0150. Print 2023 Feb 1. Reproduction. 2023. PMID: 36322478 Review.
Cited by
-
Endometrial proteomic profile of patients with repeated implantation failure.Front Endocrinol (Lausanne). 2023 Jul 31;14:1144393. doi: 10.3389/fendo.2023.1144393. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37583433 Free PMC article.
-
Screening of Diagnostic Biomarkers and Immune Infiltration Characteristics Linking Rheumatoid Arthritis and Rosacea Based on Bioinformatics Analysis.J Inflamm Res. 2024 Aug 1;17:5177-5195. doi: 10.2147/JIR.S467760. eCollection 2024. J Inflamm Res. 2024. PMID: 39104909 Free PMC article.
-
Integrated analysis of single-cell and bulk RNA sequencing data identifies the characteristics of ferroptosis in lumbar disc herniation.Funct Integr Genomics. 2023 Sep 1;23(3):289. doi: 10.1007/s10142-023-01216-8. Funct Integr Genomics. 2023. PMID: 37653201
-
Analysis of weighted gene co-expression networks and clinical validation identify hub genes and immune cell infiltration in the endometrial cells of patients with recurrent implantation failure.Front Genet. 2024 Apr 5;15:1292757. doi: 10.3389/fgene.2024.1292757. eCollection 2024. Front Genet. 2024. PMID: 38645487 Free PMC article.
-
WD-repeat containing protein-61 regulates endometrial epithelial cell adhesion indicating an important role in receptivity.Mol Hum Reprod. 2024 Nov 14;30(11):gaae039. doi: 10.1093/molehr/gaae039. Mol Hum Reprod. 2024. PMID: 39531333 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

