Feeding Asian honeybee queens with European honeybee royal jelly alters body color and expression of related coding and non-coding RNAs
- PMID: 36776963
- PMCID: PMC9908965
- DOI: 10.3389/fphys.2023.1073625
Feeding Asian honeybee queens with European honeybee royal jelly alters body color and expression of related coding and non-coding RNAs
Abstract
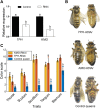
Background and aims: The Asian honeybee (Apis cerana) and the European honeybee (Apis mellifera) are reproductively isolated. Previous studies reported that exchanging the larval food between the two species, known as nutritional crossbreeding, resulted in obvious changes in morphology, physiology and behavior. This study explored the molecular mechanisms underlying the honeybee nutritional crossbreeding. Methods: This study used full nutritional crossbreeding technology to rear A. cerana queens by feeding them with an A. mellifera royal jelly-based diet in an incubator. The body color and the expression of certain genes, microRNA, lncRNA, and circRNA among nutritional crossbred A. cerana queens (NQ), and control A. cerana queens (CQ) were compared. The biological functions of two target genes, TPH1 and KMO, were verified using RNA interference. Results: Our results showed that the NQ's body color turned yellow compared to the black control queens. Whole transcriptome sequencing results showed that a total of 1484, 311, 92, and 169 DEGs, DElncRNAs, DEmiRNAs, and DEcircRNAs, respectively, were identified in NQ and CQ, in which seven DEGs were enriched for three key pathways (tryptophan, tyrosine, and dopamine) involved in melanin synthesis. Interestingly, eight DElncRNAs and three DEmiRNAs were enriched into the key pathways regulating the above key DEGs. No circRNAs were enriched into these key pathways. Knocking down two key genes (KMO and TPH1) resulted in altered body color, suggesting that feeding NQ's an RNAi-based diet significantly downregulated the expression of TPH1 and KMO in 4-day-old larvae, which confirmed the function of key DEGs in the regulation of honeybee body color. Conclusion: These findings reveal that the larval diets from A. mellifera could change the body color of A. cerana, perhaps by altering the expression of non-coding RNAs and related key genes. This study serves as a model of epigenetic regulation in insect body color induced by environmental factors.
Keywords: body color alteration; gene expression; honeybees; non-coding RNA expression; nutritional crossbreed.
Copyright © 2023 Abdelmawla, Yang, Li, Li, Li, Liu, He and Zeng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Arakane Y., Noh M. Y., Asano T., Kramer J., K. (2016). “Tyrosine metabolism for insect cuticle pigmentation and sclerotization,” in Extracellular composite matrices in arthropods. Editors Cohen E., Moussian B. (Switzerland, 156–220. 10.1007/978-3-319-40740-1_6 - DOI
-
- Chen H., Li F. M., Zheng M. J., Cao D. D., He Z. M., Yang M. X. (2015). The effect of nutritional hybridization on the reproductive potential of the queen bee in China. Sichuan Anim. Husb. Vet. Med. 42 (09), 24–26. (in Chinese).
LinkOut - more resources
Full Text Sources

