Virtual Fly Brain-An interactive atlas of the Drosophila nervous system
- PMID: 36776967
- PMCID: PMC9908962
- DOI: 10.3389/fphys.2023.1076533
Virtual Fly Brain-An interactive atlas of the Drosophila nervous system
Abstract
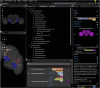
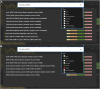
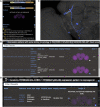
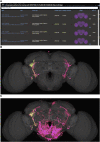
As a model organism, Drosophila is uniquely placed to contribute to our understanding of how brains control complex behavior. Not only does it have complex adaptive behaviors, but also a uniquely powerful genetic toolkit, increasingly complete dense connectomic maps of the central nervous system and a rapidly growing set of transcriptomic profiles of cell types. But this also poses a challenge: Given the massive amounts of available data, how are researchers to Find, Access, Integrate and Reuse (FAIR) relevant data in order to develop an integrated anatomical and molecular picture of circuits, inform hypothesis generation, and find reagents for experiments to test these hypotheses? The Virtual Fly Brain (virtualflybrain.org) web application & API provide a solution to this problem, using FAIR principles to integrate 3D images of neurons and brain regions, connectomics, transcriptomics and reagent expression data covering the whole CNS in both larva and adult. Users can search for neurons, neuroanatomy and reagents by name, location, or connectivity, via text search, clicking on 3D images, search-by-image, and queries by type (e.g., dopaminergic neuron) or properties (e.g., synaptic input in the antennal lobe). Returned results include cross-registered 3D images that can be explored in linked 2D and 3D browsers or downloaded under open licenses, and extensive descriptions of cell types and regions curated from the literature. These solutions are potentially extensible to cover similar atlasing and data integration challenges in vertebrates.
Keywords: FAIR; atlas; connectomics; drosophila; neurobiology; ontology; transcriptomics.
Copyright © 2023 Court, Costa, Pilgrim, Millburn, Holmes, McLachlan, Larkin, Matentzoglu, Kir, Parkinson, Brown, O’Kane, Armstrong, Jefferis and Osumi-Sutherland.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

