Using PDX animal models to identify and stratify adenoid cystic carcinoma patients presenting an enhanced response to HDAC inhibitors
- PMID: 36777521
- PMCID: PMC9906074
Using PDX animal models to identify and stratify adenoid cystic carcinoma patients presenting an enhanced response to HDAC inhibitors
Abstract
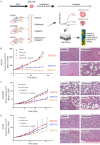
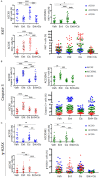
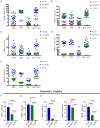
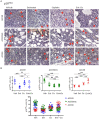
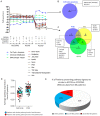
Adenoid cystic carcinoma (ACC) patients face a highly infiltrative and metastatic disease characterized by poor survival rates and suboptimal response to available therapies. We have previously shown that sensitization of ACC tumors to chemotherapy using histone deacetylase inhibitors (HDACi) constitutes a promising therapeutic strategy to manage tumor growth. Here, we used patient-derived xenografts (PDX) from ACC tumors to evaluate the effects of in vivo administration of the HDAC inhibitor Entinostat combined with Cisplatin over tumor growth. RNA from PDX tumor samples receiving the proposed therapy were analyzed using NanoString technology to identify molecular signatures capable of predicting ACC response to the therapy. We also used an RNAseq dataset from 68 ACC patients to validate the molecular signature identified by the NanoString platform. We found that the administration of Entinostat combined with Cisplatin resulted in a potent tumor growth inhibition (TGI) ranging from 38% to 106% of the original tumor mass. Enhanced response to therapy is consistent with the reactivation of tumor suppressor genes, including SFRP1, and the downregulation of oncogenes like FGF8 and CCR7. Nanostring data from PDX tumors identified a genetic signature capable of predicting tumor response to therapy. We further stratified 68 ACC patients containing RNAseq data accordingly to the activity levels of the identified genetic signature. We found that 23% of all patients exhibit a genetic signature consistent with a high ACC tumor response rate to Entinostat and Cisplatin. Our study provides compelling preclinical data supporting the deployment of a powerful systemic anticancer therapy crafted and explicitly tested for ACC tumors.
Keywords: Precision medicine; acetylation; histone; senescence; tumor genome landscape.
AJCR Copyright © 2023.
Conflict of interest statement
None.
Figures

References
-
- Xu B, Drill E, Ho A, Ho A, Dunn L, Prieto-Granada CN, Chan T, Ganly I, Ghossein R, Katabi N. Predictors of outcome in adenoid cystic carcinoma of salivary glands: a clinicopathologic study with correlation between MYB fusion and protein expression. Am J Surg Pathol. 2017;41:1422–1432. - PMC - PubMed
-
- Papaspyrou G, Hoch S, Rinaldo A, Rodrigo JP, Takes RP, van Herpen C, Werner JA, Ferlito A. Chemotherapy and targeted therapy in adenoid cystic carcinoma of the head and neck: a review. Head Neck. 2011;33:905–911. - PubMed
-
- Ho AS, Ochoa A, Jayakumaran G, Zehir A, Valero Mayor C, Tepe J, Makarov V, Dalin MG, He J, Bailey M, Montesion M, Ross JS, Miller VA, Chan L, Ganly I, Dogan S, Katabi N, Tsipouras P, Ha P, Agrawal N, Solit DB, Futreal PA, El Naggar AK, Reis-Filho JS, Weigelt B, Ho AL, Schultz N, Chan TA, Morris LG. Genetic hallmarks of recurrent/metastatic adenoid cystic carcinoma. J Clin Invest. 2019;129:4276–4289. - PMC - PubMed
LinkOut - more resources
Full Text Sources
