Systematic genetic analysis of pediatric patients with autoinflammatory diseases
- PMID: 36777733
- PMCID: PMC9911692
- DOI: 10.3389/fgene.2023.1065907
Systematic genetic analysis of pediatric patients with autoinflammatory diseases
Abstract
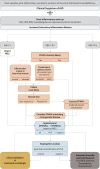
Monogenic autoinflammatory diseases (AID) encompass a growing group of inborn errors of the innate immune system causing unprovoked or exaggerated systemic inflammation. Diagnosis of monogenic AID requires an accurate description of the patients' phenotype, and the identification of highly penetrant genetic variants in single genes is pivotal. We performed whole exome sequencing (WES) of 125 pediatric patients with suspected monogenic AID in a routine genetic diagnostic setting. Datasets were analyzed in a step-wise approach to identify the most feasible diagnostic strategy. First, we analyzed a virtual gene panel including 13 genes associated with known AID and, if no genetic diagnosis was established, we then analyzed a virtual panel including 542 genes published by the International Union of Immunological Societies associated including all known inborn error of immunity (IEI). Subsequently, WES data was analyzed without pre-filtering for known AID/IEI genes. Analyzing 13 genes yielded a definite diagnosis in 16.0% (n = 20). The diagnostic yield was increased by analyzing 542 genes to 20.8% (n = 26). Importantly, expanding the analysis to WES data did not increase the diagnostic yield in our cohort, neither in single WES analysis, nor in trio-WES analysis. The study highlights that the cost- and time-saving analysis of virtual gene panels is sufficient to rapidly confirm the differential diagnosis in pediatric patients with AID. WES data or trio-WES data analysis as a first-tier diagnostic analysis in patients with suspected monogenic AID is of limited benefit.
Keywords: FMF; autoinflammatory diseases; genetic diagnostics; inborn errors of immunity (IEI); whole exome sequencing (WES).
Copyright © 2023 Poker, von Hardenberg, Hofmann, Tang, Baumann, Schwerk, Wetzke, Lindenthal, Auber, Schlegelberger, Ott, von Bismarck, Viemann, Dressler, Klemann and Bergmann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Trio-based whole exome sequencing in patients with suspected sporadic inborn errors of immunity: A retrospective cohort study.Elife. 2022 Oct 17;11:e78469. doi: 10.7554/eLife.78469. Elife. 2022. PMID: 36250618 Free PMC article.
-
Assessing whole-exome sequencing data from undiagnosed Brazilian patients to improve the diagnostic yield of inborn errors of immunity.BMC Genom Data. 2023 Jun 30;24(1):36. doi: 10.1186/s12863-023-01137-2. BMC Genom Data. 2023. PMID: 37391719 Free PMC article.
-
Genetic screening in a Brazilian cohort with inborn errors of immunity.BMC Genom Data. 2023 Aug 17;24(1):47. doi: 10.1186/s12863-023-01148-z. BMC Genom Data. 2023. PMID: 37592284 Free PMC article.
-
Current genetic diagnostics in inborn errors of immunity.Front Pediatr. 2024 Apr 10;12:1279112. doi: 10.3389/fped.2024.1279112. eCollection 2024. Front Pediatr. 2024. PMID: 38659694 Free PMC article. Review.
-
Effect of Whole Exome Sequencing in Diagnosis of Inborn Errors of Metabolism and Neurogenetic Disorders.Iran J Child Neurol. 2018 Winter;12(1):7-15. Iran J Child Neurol. 2018. PMID: 29379558 Free PMC article. Review.
Cited by
-
Development and internal validation of a clinical and genetic risk score for rheumatoid arthritis-associated interstitial lung disease.Rheumatology (Oxford). 2025 Jan 1;64(1):268-275. doi: 10.1093/rheumatology/keae001. Rheumatology (Oxford). 2025. PMID: 38243706 Free PMC article.
-
Consanguinity and rare monogenic systemic autoinflammatory disorders: implications for prevalence and genetic variability.Pediatr Rheumatol Online J. 2025 Jul 30;23(1):83. doi: 10.1186/s12969-025-01133-z. Pediatr Rheumatol Online J. 2025. PMID: 40739506 Free PMC article.
-
Unravelling the clinical heterogeneity of undefined recurrent fever over time in the European registries on Autoinflammation.Pediatr Rheumatol Online J. 2024 May 17;22(1):55. doi: 10.1186/s12969-024-00987-z. Pediatr Rheumatol Online J. 2024. PMID: 38760816 Free PMC article.
References
-
- Basel-Salmon L., Orenstein N., Markus-Bustani K., Ruhrman-Shahar N., Kilim Y., Magal N., et al. (2019). Improved diagnostics by exome sequencing following raw data reevaluation by clinical geneticists involved in the medical care of the individuals tested. Genet. Med. 21 (6), 1443–1451. 10.1038/s41436-018-0343-7 - DOI - PubMed
LinkOut - more resources
Full Text Sources

