Type 1 diabetes risk genes mediate pancreatic beta cell survival in response to proinflammatory cytokines
- PMID: 36778047
- PMCID: PMC9903835
- DOI: 10.1016/j.xgen.2022.100214
Type 1 diabetes risk genes mediate pancreatic beta cell survival in response to proinflammatory cytokines
Abstract
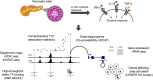
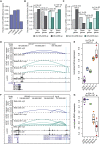
We combined functional genomics and human genetics to investigate processes that affect type 1 diabetes (T1D) risk by mediating beta cell survival in response to proinflammatory cytokines. We mapped 38,931 cytokine-responsive candidate cis-regulatory elements (cCREs) in beta cells using ATAC-seq and snATAC-seq and linked them to target genes using co-accessibility and HiChIP. Using a genome-wide CRISPR screen in EndoC-βH1 cells, we identified 867 genes affecting cytokine-induced survival, and genes promoting survival and up-regulated in cytokines were enriched at T1D risk loci. Using SNP-SELEX, we identified 2,229 variants in cytokine-responsive cCREs altering transcription factor (TF) binding, and variants altering binding of TFs regulating stress, inflammation, and apoptosis were enriched for T1D risk. At the 16p13 locus, a fine-mapped T1D variant altering TF binding in a cytokine-induced cCRE interacted with SOCS1, which promoted survival in cytokine exposure. Our findings reveal processes and genes acting in beta cells during inflammation that modulate T1D risk.
Keywords: 3D chromatin interactions; CRISPR screen; accessible chromatin; beta cell; functional genomics; gene expression; high-throughput reporter assay; human genetics; proinflammatory cytokines; type 1 diabetes.
© 2022.
Conflict of interest statement
K.J.G. is a consultant of Genentech and holds stock in Neurocrine Biosciences. B.R. is a consultant of Arima Genomics and a co-founder of Epigenome Technologies. P.B. is an employee of Shoreline Bioscience. N.N. is an employee of Guardant Health. E.B. is an employee and shareholder of Aetion. K.K. is an employee of Cartography Bio. Y.Q. is an employee of Sana Biotechnology. M.D. is an employee and shareholder of Seer. J.C. is an employee and shareholder of Pfizer.
Figures





References
-
- Brozzi F., Nardelli T.R., Lopes M., Millard I., Barthson J., Igoillo-Esteve M., Grieco F.A., Villate O., Oliveira J.M., Casimir M., et al. Cytokines induce endoplasmic reticulum stress in human, rat and mouse beta cells via different mechanisms. Diabetologia. 2015;58:2307–2316. doi: 10.1007/s00125-015-3669-6. - DOI - PubMed
-
- Ortis F., Naamane N., Flamez D., Ladrière L., Moore F., Cunha D.A., Colli M.L., Thykjaer T., Thorsen K., Orntoft T.F., Eizirik D.L. Cytokines interleukin-1 and tumor necrosis factor- regulate different transcriptional and alternative splicing networks in primary -cells. Diabetes. 2010;59:358–374. doi: 10.2337/db09-1159. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

