Targeted long-read sequencing facilitates phased diploid assembly and genotyping of the human T cell receptor alpha, delta, and beta loci
- PMID: 36778049
- PMCID: PMC9903726
- DOI: 10.1016/j.xgen.2022.100228
Targeted long-read sequencing facilitates phased diploid assembly and genotyping of the human T cell receptor alpha, delta, and beta loci
Abstract
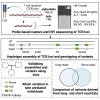
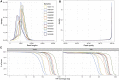
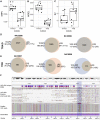
T cell receptors (TCRs) recognize peptide fragments presented by the major histocompatibility complex (MHC) and are critical to T cell-mediated immunity. Recent data have indicated that genetic diversity within TCR-encoding gene regions is underexplored, limiting understanding of the impact of TCR loci polymorphisms on TCR function in disease, even though TCR repertoire signatures (1) are heritable and (2) associate with disease phenotypes. To address this, we developed a targeted long-read sequencing approach to generate highly accurate haplotype resolved assemblies of the TCR beta (TRB) and alpha/delta (TRA/D) loci, facilitating the genotyping of all variant types, including structural variants. We validate our approach using two mother-father-child trios and 5 unrelated donors representing multiple populations. This resulted in improved genotyping accuracy and the discovery of 84 undocumented V, D, J, and C alleles, demonstrating the utility of this framework for improving our understanding of TCR diversity and function in disease.
Keywords: T cell receptor; TCR; TCR alleles; alpha; beta; delta; immunogenomics; long-read sequencing; long-reads; structural variants.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
T cell receptor beta germline variability is revealed by inference from repertoire data.Genome Med. 2022 Jan 7;14(1):2. doi: 10.1186/s13073-021-01008-4. Genome Med. 2022. PMID: 34991709 Free PMC article.
-
A new high-throughput sequencing method for determining diversity and similarity of T cell receptor (TCR) α and β repertoires and identifying potential new invariant TCR α chains.BMC Immunol. 2016 Oct 11;17(1):38. doi: 10.1186/s12865-016-0177-5. BMC Immunol. 2016. PMID: 27729009 Free PMC article.
-
Ultrasensitive allele inference from immune repertoire sequencing data with MiXCR.bioRxiv [Preprint]. 2023 Nov 17:2023.10.10.561703. doi: 10.1101/2023.10.10.561703. bioRxiv. 2023. Update in: Genome Res. 2024 Dec 23;34(12):2293-2303. doi: 10.1101/gr.278775.123. PMID: 38014266 Free PMC article. Updated. Preprint.
-
Haplotyping-Assisted Diploid Assembly and Variant Detection with Linked Reads.Methods Mol Biol. 2023;2590:161-182. doi: 10.1007/978-1-0716-2819-5_11. Methods Mol Biol. 2023. PMID: 36335499 Review.
-
Repertoire development and ligand specificity of murine TCR gamma delta cells.Immunol Rev. 1991 Apr;120:5-33. doi: 10.1111/j.1600-065x.1991.tb00585.x. Immunol Rev. 1991. PMID: 1650760 Review.
Cited by
-
The human immunoglobulin heavy chain constant gene locus is enriched for large complex structural variants and coding polymorphisms that vary in frequency among human populations.bioRxiv [Preprint]. 2025 Feb 12:2025.02.12.634878. doi: 10.1101/2025.02.12.634878. bioRxiv. 2025. PMID: 39990387 Free PMC article. Preprint.
-
Digger: directed annotation of immunoglobulin and T cell receptor V, D, and J gene sequences and assemblies.Bioinformatics. 2024 Mar 4;40(3):btae144. doi: 10.1093/bioinformatics/btae144. Bioinformatics. 2024. PMID: 38478393 Free PMC article.
-
Ultrasensitive allele inference from immune repertoire sequencing data with MiXCR.Genome Res. 2024 Dec 23;34(12):2293-2303. doi: 10.1101/gr.278775.123. Genome Res. 2024. PMID: 39433438 Free PMC article.
-
AIRR community curation and standardised representation for immunoglobulin and T cell receptor germline sets.Immunoinformatics (Amst). 2023 Jun;10:100025. doi: 10.1016/j.immuno.2023.100025. Epub 2023 Feb 19. Immunoinformatics (Amst). 2023. PMID: 37388275 Free PMC article.
-
A template for multitudes: Germline immune polymorphism of the T cell receptor loci.Cell Genom. 2022 Dec 14;2(12):100231. doi: 10.1016/j.xgen.2022.100231. eCollection 2022 Dec 14. Cell Genom. 2022. PMID: 36778048 Free PMC article.
References
-
- Lefranc M.-P., Lefranc G. Academic Press; 2001. The T Cell Receptor FactsBook.
-
- Nikolich-Žugich J., Slifka M.K., Messaoudi I. The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 2004;4:123–132. - PubMed
-
- Zvyagin I.V., Pogorelyy M.V., Ivanova M.E., Komech E.A., Shugay M., Bolotin D.A., Shelenkov A.A., Kurnosov A.A., Staroverov D.B., Chudakov D.M., et al. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc. Natl. Acad. Sci. USA. 2014;111:5980–5985. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

