Multi-ancestry meta-analysis of asthma identifies novel associations and highlights the value of increased power and diversity
- PMID: 36778051
- PMCID: PMC9903683
- DOI: 10.1016/j.xgen.2022.100212
Multi-ancestry meta-analysis of asthma identifies novel associations and highlights the value of increased power and diversity
Abstract
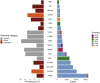
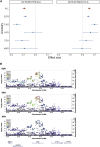
Asthma is a complex disease that varies widely in prevalence across populations. The extent to which genetic variation contributes to these disparities is unclear, as the genetics underlying asthma have been investigated primarily in populations of European descent. As part of the Global Biobank Meta-analysis Initiative, we conducted a large-scale genome-wide association study of asthma (153,763 cases and 1,647,022 controls) via meta-analysis across 22 biobanks spanning multiple ancestries. We discovered 179 asthma-associated loci, 49 of which were not previously reported. Despite the wide range in asthma prevalence among biobanks, we found largely consistent genetic effects across biobanks and ancestries. The meta-analysis also improved polygenic risk prediction in non-European populations compared with previous studies. Additionally, we found considerable genetic overlap between age-of-onset subtypes and between asthma and comorbid diseases. Our work underscores the multi-factorial nature of asthma development and offers insight into its shared genetic architecture.
Keywords: GWAS; asthma; cross-trait; heterogeneity; meta-analysis; multi-ancestry; polygenic risk prediction; subtypes.
© 2022 The Author(s).
Conflict of interest statement
M.J.D. is a founder of Maze Therapeutics. B.M.N. is a member of the scientific advisory board at Deep Genomics and consultant for Camp4 Therapeutics, Takeda Pharmaceutical, and Biogen.
Figures




References
-
- Moffatt M.F., Gut I.G., Demenais F., Strachan D.P., Bouzigon E., Heath S., von Mutius E., Farrall M., Lathrop M., Cookson W.O.C.M., GABRIEL Consortium A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

