ESRG is critical to maintain the cell survival and self-renewal/pluripotency of hPSCs by collaborating with MCM2 to suppress p53 pathway
- PMID: 36778110
- PMCID: PMC9909993
- DOI: 10.7150/ijbs.79095
ESRG is critical to maintain the cell survival and self-renewal/pluripotency of hPSCs by collaborating with MCM2 to suppress p53 pathway
Abstract
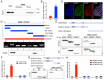
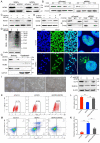
The mechanisms of self-renewal and pluripotency maintenance of human pluripotent stem cells (hPSCs) have not been fully elucidated, especially for the role of those poorly characterized long noncoding RNAs (lncRNAs). ESRG is a lncRNA highly expressed in hPSCs, and its functional roles are being extensively explored in the field. Here, we identified that the transcription of ESRG can be directly regulated by OCT4, a key self-renewal factor in hPSCs. Knockdown of ESRG induces hPSC differentiation, cell cycle arrest, and apoptosis. ESRG binds to MCM2, a replication-licensing factor, to sustain its steady-state level and nuclear location, safeguarding error-free DNA replication. Further study showed that ESRG knockdown leads to MCM2 abnormalities, resulting in DNA damage and activation of the p53 pathway, ultimately impairs hPSC self-renewal and pluripotency, and induces cell apoptosis. In summary, our study suggests that ESRG, as a novel target of OCT4, plays an essential role in maintaining the cell survival and self-renewal/pluripotency of hPSCs in collaboration with MCM2 to suppress p53 signaling. These findings provide critical insights into the mechanisms underlying the maintenance of self-renewal and pluripotency in hPSCs by lncRNAs.
Keywords: ESRG; MCM2; OCT4; cell survival; human pluripotent stem cells; p53; pluripotency.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





Similar articles
-
Embryonic stem cell related gene regulates alternative splicing of transcription factor 3 to maintain human embryonic stem cells' self-renewal and pluripotency.Stem Cells. 2024 Jun 14;42(6):540-553. doi: 10.1093/stmcls/sxae020. Stem Cells. 2024. PMID: 38393342
-
Progesterone Receptor Membrane Component 1 suppresses the p53 and Wnt/β-catenin pathways to promote human pluripotent stem cell self-renewal.Sci Rep. 2018 Feb 14;8(1):3048. doi: 10.1038/s41598-018-21322-z. Sci Rep. 2018. PMID: 29445107 Free PMC article.
-
The pluripotent stem cell-specific transcript ESRG is dispensable for human pluripotency.PLoS Genet. 2021 May 25;17(5):e1009587. doi: 10.1371/journal.pgen.1009587. eCollection 2021 May. PLoS Genet. 2021. PMID: 34033652 Free PMC article.
-
Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells.Stem Cells Dev. 2013 Aug 15;22(16):2240-53. doi: 10.1089/scd.2013.0014. Epub 2013 May 14. Stem Cells Dev. 2013. PMID: 23528033 Free PMC article. Review.
-
Long noncoding RNAs in cell differentiation and pluripotency.Cell Tissue Res. 2016 Dec;366(3):509-521. doi: 10.1007/s00441-016-2451-5. Epub 2016 Jun 30. Cell Tissue Res. 2016. PMID: 27365087 Review.
Cited by
-
Single-cell transcriptomics reveals the molecular basis of human iPS cell differentiation into ectodermal ocular lineages.Commun Biol. 2024 Nov 12;7(1):1495. doi: 10.1038/s42003-024-07130-4. Commun Biol. 2024. PMID: 39532995 Free PMC article.
-
Towards Stem/Progenitor Cell-Based Therapies for Retinal Degeneration.Stem Cell Rev Rep. 2024 Aug;20(6):1459-1479. doi: 10.1007/s12015-024-10740-4. Epub 2024 May 29. Stem Cell Rev Rep. 2024. PMID: 38809490 Review.
-
Construction of functional neural network tissue combining CBD-NT3-modified linear-ordered collagen scaffold and TrkC-modified iPSC-derived neural stem cells for spinal cord injury repair.Bioact Mater. 2024 Feb 3;35:242-258. doi: 10.1016/j.bioactmat.2024.01.012. eCollection 2024 May. Bioact Mater. 2024. PMID: 38333615 Free PMC article.
-
Hydrogen sulfide preserves intestinal barrier repair function through sulfhydration of RPS20 in experimental colitis.Sci Rep. 2025 May 21;15(1):17673. doi: 10.1038/s41598-025-02268-5. Sci Rep. 2025. PMID: 40399407 Free PMC article.
-
The role of non-coding RNA regulates stem cell programmed death in disease therapy.Noncoding RNA Res. 2025 Apr 23;13:57-70. doi: 10.1016/j.ncrna.2025.04.005. eCollection 2025 Aug. Noncoding RNA Res. 2025. PMID: 40454168 Free PMC article. Review.
References
-
- Thomson JA, Itskovitz-Eldor J, Shapiro SS. et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. - PubMed
-
- Wang Z, Oron E, Nelson B. et al. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

