This is a preprint.
Archaic Introgression Shaped Human Circadian Traits
- PMID: 36778254
- PMCID: PMC9915721
- DOI: 10.1101/2023.02.03.527061
Archaic Introgression Shaped Human Circadian Traits
Update in
-
Archaic Introgression Shaped Human Circadian Traits.Genome Biol Evol. 2023 Dec 1;15(12):evad203. doi: 10.1093/gbe/evad203. Genome Biol Evol. 2023. PMID: 38095367 Free PMC article.
Abstract
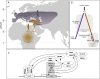
Introduction: When the ancestors of modern Eurasians migrated out of Africa and interbred with Eurasian archaic hominins, namely Neanderthals and Denisovans, DNA of archaic ancestry integrated into the genomes of anatomically modern humans. This process potentially accelerated adaptation to Eurasian environmental factors, including reduced ultra-violet radiation and increased variation in seasonal dynamics. However, whether these groups differed substantially in circadian biology, and whether archaic introgression adaptively contributed to human chronotypes remains unknown.
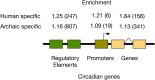
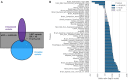
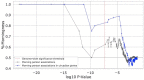
Results: Here we traced the evolution of chronotype based on genomes from archaic hominins and present-day humans. First, we inferred differences in circadian gene sequences, splicing, and regulation between archaic hominins and modern humans. We identified 28 circadian genes containing variants with potential to alter splicing in archaics (e.g., CLOCK, PER2, RORB, RORC), and 16 circadian genes likely divergently regulated between present-day humans and archaic hominins, including RORA. These differences suggest the potential for introgression to modify circadian gene expression. Testing this hypothesis, we found that introgressed variants are enriched among eQTLs for circadian genes. Supporting the functional relevance of these regulatory effects, we found that many introgressed alleles have associations with chronotype. Strikingly, the strongest introgressed effects on chronotype increase morningness, consistent with adaptations to high latitude in other species. Finally, we identified several circadian loci with evidence of adaptive introgression or latitudinal clines in allele frequency.
Conclusions: These findings identify differences in circadian gene regulation between modern humans and archaic hominins and support the contribution of introgression via coordinated effects on variation in human chronotype.
Keywords: Neanderthals; adaptive evolution; adaptive introgression; chronotype; circadian biology; gene expression.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare that they have no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
