Nuclear translocation of spike mRNA and protein is a novel feature of SARS-CoV-2
- PMID: 36778849
- PMCID: PMC9909199
- DOI: 10.3389/fmicb.2023.1073789
Nuclear translocation of spike mRNA and protein is a novel feature of SARS-CoV-2
Abstract
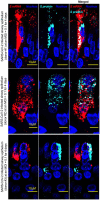
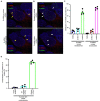
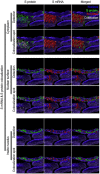
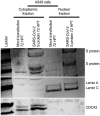
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes severe pathophysiology in vulnerable older populations and appears to be highly pathogenic and more transmissible than other coronaviruses. The spike (S) protein appears to be a major pathogenic factor that contributes to the unique pathogenesis of SARS-CoV-2. Although the S protein is a surface transmembrane type 1 glycoprotein, it has been predicted to be translocated into the nucleus due to the novel nuclear localization signal (NLS) "PRRARSV," which is absent from the S protein of other coronaviruses. Indeed, S proteins translocate into the nucleus in SARS-CoV-2-infected cells. S mRNAs also translocate into the nucleus. S mRNA colocalizes with S protein, aiding the nuclear translocation of S mRNA. While nuclear translocation of nucleoprotein (N) has been shown in many coronaviruses, the nuclear translocation of both S mRNA and S protein reveals a novel feature of SARS-CoV-2.
Keywords: NLS; SARS-CoV-2; mRNA; nuclear translocation; spike.
Copyright © 2023 Sattar, Kabat, Jerome, Feldmann, Bailey and Mehedi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Update of
-
Nuclear translocation of spike mRNA and protein is a novel pathogenic feature of SARS-CoV-2.bioRxiv [Preprint]. 2022 Sep 27:2022.09.27.509633. doi: 10.1101/2022.09.27.509633. bioRxiv. 2022. Update in: Front Microbiol. 2023 Jan 26;14:1073789. doi: 10.3389/fmicb.2023.1073789. PMID: 36203551 Free PMC article. Updated. Preprint.
Similar articles
-
Nuclear translocation of spike mRNA and protein is a novel pathogenic feature of SARS-CoV-2.bioRxiv [Preprint]. 2022 Sep 27:2022.09.27.509633. doi: 10.1101/2022.09.27.509633. bioRxiv. 2022. Update in: Front Microbiol. 2023 Jan 26;14:1073789. doi: 10.3389/fmicb.2023.1073789. PMID: 36203551 Free PMC article. Updated. Preprint.
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
-
The human pandemic coronaviruses on the show: The spike glycoprotein as the main actor in the coronaviruses play.Int J Biol Macromol. 2021 May 15;179:1-19. doi: 10.1016/j.ijbiomac.2021.02.203. Epub 2021 Mar 2. Int J Biol Macromol. 2021. PMID: 33667553 Free PMC article. Review.
-
UBR5 Acts as an Antiviral Host Factor against MERS-CoV via Promoting Ubiquitination and Degradation of ORF4b.J Virol. 2022 Sep 14;96(17):e0074122. doi: 10.1128/jvi.00741-22. Epub 2022 Aug 18. J Virol. 2022. PMID: 35980206 Free PMC article.
-
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Membrane (M) and Spike (S) Proteins Antagonize Host Type I Interferon Response.Front Cell Infect Microbiol. 2021 Dec 7;11:766922. doi: 10.3389/fcimb.2021.766922. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34950606 Free PMC article.
Cited by
-
Unveiling the native architecture of adult cardiac tissue using the 3D-NaissI method.Cell Mol Life Sci. 2025 Feb 5;82(1):70. doi: 10.1007/s00018-025-05595-y. Cell Mol Life Sci. 2025. PMID: 39907789 Free PMC article.
-
Pro Inflammatory Cytokines Profiles of Patients With Long COVID Differ Between Variant Epochs.J Prim Care Community Health. 2024 Jan-Dec;15:21501319241254751. doi: 10.1177/21501319241254751. J Prim Care Community Health. 2024. PMID: 38808863 Free PMC article.
-
Significant Increase in Excess Deaths after Repeated COVID-19 Vaccination in Japan.JMA J. 2025 Apr 28;8(2):584-586. doi: 10.31662/jmaj.2024-0298. Epub 2025 Mar 7. JMA J. 2025. PMID: 40416011 Free PMC article.
-
mRNA: Vaccine or Gene Therapy? The Safety Regulatory Issues.Int J Mol Sci. 2023 Jun 22;24(13):10514. doi: 10.3390/ijms241310514. Int J Mol Sci. 2023. PMID: 37445690 Free PMC article. Review.
-
Safety of COVID-19 Vaccines in Patients with Autoimmune Diseases, in Patients with Cardiac Issues, and in the Healthy Population.Pathogens. 2023 Feb 2;12(2):233. doi: 10.3390/pathogens12020233. Pathogens. 2023. PMID: 36839505 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

